Abstract
Purpose
To study the application of next-generation sequencing on preimplantation genetic testing for recurrent hydatidiform mole patients.
Methods
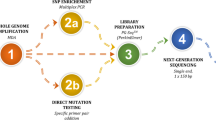
A total of ten recurrent hydatidiform mole patients aged 27–34 years with a history of at least twice hydatidiform moles and no normal pregnancy were collected from 2019 to 2020. The diagnosis of hydatidiform mole type was clarified using short tandem repeat genotyping on products of conception, and whole-exome sequencing was applied for all patients and their partners. Seven recurrent hydatidiform mole patients with complete hydatidiform mole/partial hydatidiform mole type among previous hydatidiform mole tissues and no Pathogenetic/Likely pathogenetic/Uncertain significance variants in NLRP7/KHDC3L/MEI1/C11orf80 underwent a procedure of preimplantation genetic testing. Next-generation sequencing for analyzing the copy number variants and the numbers of heterozygous single nucleotide polymorphism was adopted to clarify the ploidy and parental origin of the embryo chromosomes in vitro. Embryos with biparental diploidy were selected for transfer.
Results
Seven patients have undergone the procedure of preimplantation genetic testing, and twenty-three embryos were obtained, among which 82.6% (n = 19) were identified transferrable and 17.4% (n = 4) were identified aneuploid. Two patients have delivered healthy babies and another is currently in the second trimester after transfer.
Conclusion
Analyzing the copy number variants and the numbers of heterozygous single nucleotide polymorphism on the basis of next-generation sequencing can be utilized in the procedure of preimplantation genetic testing among part of recurrent hydatidiform mole patients. The current study is effective to reduce the occurrence of hydatidiform mole with improved clinical strategy, the advanced testing technology and analysis methods, as three of seven patients have conceived or delivered successfully.





Similar content being viewed by others
References
Heller DS. Update on the pathology of gestational trophoblastic disease. APMIS. 2018;126(7):647–54.
Ronnett BM. Hydatidiform Moles: ancillary techniques to refine diagnosis. Arch Pathol Lab Med. 2018;142(12):1485–502.
Kalogiannidis I, et al. Recurrent complete hydatidiform mole: where we are, is there a safe gestational horizon? Opinion and mini-review. J Assist Reprod Genet. 2018;35(6):967–73.
Braga A, et al. Challenges in the diagnosis and treatment of gestational trophoblastic neoplasia worldwide. World J Clin Oncol. 2019;10(2):28–37.
Moein-Vaziri N, et al. Clinical and genetic-epigenetic aspects of recurrent hydatidiform mole: a review of literature. Taiwan J Obstet Gynecol. 2018;57(1):1–6.
Alici-Garipcan A, et al. NLRP7 plays a functional role in regulating BMP4 signaling during differentiation of patient-derived trophoblasts. Cell Death Dis. 2020;11(8):658.
Demond H, et al. A KHDC3L mutation resulting in recurrent hydatidiform mole causes genome-wide DNA methylation loss in oocytes and persistent imprinting defects post-fertilisation. Genome Med. 2019;11(1):84.
Messaed C, et al. NLRP7 in the spectrum of reproductive wastage: rare non-synonymous variants confer genetic susceptibility to recurrent reproductive wastage. J Med Genet. 2011;48(8):540–8.
Rezaei M, et al. Two novel mutations in the KHDC3L gene in Asian patients with recurrent hydatidiform mole. Hum Genome Var. 2016;3:16027.
Wang X, et al. Novel mutations in genes encoding subcortical maternal complex proteins may cause human embryonic developmental arrest. Reprod Biomed Online. 2018;36(6):698–704.
Pal L, et al. High incidence of triploidy in in-vitro fertilized oocytes from a patient with a previous history of recurrent gestational trophoblastic disease. Hum Reprod. 1996;11(7):1529–32.
Group, E.P.-S.P.-A.W., et al. ESHRE PGT Consortium good practice recommendations for the detection of structural and numerical chromosomal aberrations. Hum Reprod Open. 2020. 2020(3): hoaa017.
Med ASR, Technology SAR. The use of preimplantation genetic testing for aneuploidy (PGT-A): a committee opinion. Fertil Steril. 2018;109(3):429–36.
Reubinoff BE, et al. Intracytoplasmic sperm injection combined with preimplantation genetic diagnosis for the prevention of recurrent gestational trophoblastic disease. Hum Reprod. 1997;12(4):805–8.
Ogilvie CM, et al. First use of preimplantation genotyping in prevention of recurrent diandric complete hydatidiform mole. Reprod Biomed Online. 2009;19(2):224–7.
Sills ES, et al. Pathogenic variant in NLRP7 (19q13.42) associated with recurrent gestational trophoblastic disease: data from early embryo development observed during in vitro fertilization. Clin Exp Reprod Med. 2017;44(1):40–6.
Wang CM, et al. Identification of 13 novel NLRP7 mutations in 20 families with recurrent hydatidiform mole; missense mutations cluster in the leucine-rich region. J Med Genet. 2009;46(8):569–75.
Nguyen NMP, et al. The genetics of recurrent hydatidiform moles: new insights and lessons from a comprehensive analysis of 113 patients. Mod Pathol. 2018;31(7):1116–30.
Group, E.P.-M.W. et al. ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders. Hum Reprod Open. 2020;2020(3): hoaa018.
Wiszniewska J, et al. Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur J Hum Genet. 2014;22(1):79–87.
Funding
This work was supported by National Natural Science Foundation of China (81971440, 81671458) and the National Key Research and Development Program of China (2018YFC1002302).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10815_2021_2325_MOESM2_ESM.pdf
Supplementary file2 Heterozygous SNP numbers analysis for the embryos of patients RHM1 (a), RHM2 (b), RHM3 (c), RHM5 (d) and RHM6 (e). The ratio is calculated as the numbers of heterozygous SNP of maternal/paternal versus embryo on all autosomes and chromosome X. (PDF 1580 KB)
Rights and permissions
About this article
Cite this article
Yang, J., Yan, Z., Liu, Y. et al. Application of next-generation sequencing to preimplantation genetic testing for recurrent hydatidiform mole patients. J Assist Reprod Genet 38, 2881–2891 (2021). https://doi.org/10.1007/s10815-021-02325-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-021-02325-8