Abstract
Purpose
We aim to explore if there are any other candidate genetic variants in patients with a history of at least one hydatidiform mole (HM) besides the well-known variants in NLRP7 and KHDC3L.
Methods
The diagnosis of HM type was based on histopathology, and available HM tissues were collected for short tandem repeat (STR) genotyping to verify the diagnosis. DNA extracted from blood samples or decidual tissues of the 78 patients was subjected to whole-exome sequencing (WES).
Results
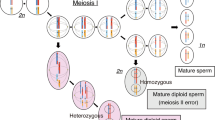
We identified five novel variants in NLRP7, two novel variants in KHDC3L, and a chromosome abnormality covering the KHDC3L locus among patients with HM. We found that patients with HM who carried heterozygous variants in KHDC3L had a chance of normal pregnancy. We also detected four novel genetic variants in candidate genes that may be associated with HM.
Conclusion
Our study enriched the spectrum of variants in NLRP7 and KHDC3L in Chinese HM patients and provided a new outlook on the effects of heterozygous variants in KHDC3L. The novel candidate genetic variants associated with HMs reported in this study will also contribute to further research on HMs.




Similar content being viewed by others
References
Khawajkie Y, et al. Comprehensive analysis of 204 sporadic hydatidiform moles: revisiting risk factors and their correlations with the molar genotypes. Mod Pathol. 2020;33(5):880–92.
Kalogiannidis I, et al. Recurrent complete hydatidiform mole: where we are, is there a safe gestational horizon? Opinion and mini-review. J Assist Reprod Genet. 2018;35(6):967–73.
Ronnett BM. Hydatidiform moles: ancillary techniques to refine diagnosis. Arch Pathol Lab Med. 2018;142(12):1485–502.
Stampone E, et al. Genetic and epigenetic control of CDKN1C expression: importance in cell commitment and differentiation, tissue homeostasis and human diseases. Int J Mol Sci. 2018;19(4):1055.
Wyner N, et al. Forensic autosomal short tandem repeats and their potential association with phenotype. Front Genet. 2020;11:884.
Bell KA, et al. Molecular genetic testing from paraffin-embedded tissue distinguishes nonmolar hydropic abortion from hydatidiform mole. Mol Diagn. 1999;4(1):11–9.
Amoushahi M, et al. The pivotal roles of the NOD-like receptors with a PYD domain, NLRPs, in oocytes and early embryo development. Biol Reprod. 2019;101(2):284–96.
Bebbere D, et al. The subcortical maternal complex: emerging roles and novel perspectives. Mol Hum Reprod. 2021;27(7):gaab043.
Moein-Vaziri N, et al. Clinical and genetic-epigenetic aspects of recurrent hydatidiform mole: a review of literature. Taiwan J Obstet Gynecol. 2018;57(1):1–6.
Fallahi J, et al. Founder effect of KHDC3L, p.M1V mutation, on Iranian patients with recurrent hydatidiform moles. Iran J Med Sci. 2020;45(2):118–124.
Wang X, et al. Novel mutations in genes encoding subcortical maternal complex proteins may cause human embryonic developmental arrest. Reprod Biomed Online. 2018;36(6):698–704.
Nguyen NMP, et al. Causative mutations and mechanism of androgenetic hydatidiform moles. Am J Hum Genet. 2018;103(5):740–51.
Qian J, et al. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur J Hum Genet. 2018;26(7):1007–13.
Zhao W, et al. Absence of KHDC3L mutations in Chinese patients with recurrent and sporadic hydatidiform moles. Cancer Genet. 2013;206(9–10):327–9.
Ji M, et al. NLRP7 and KHDC3L variants in Chinese patients with recurrent hydatidiform moles. Jpn J Clin Oncol. 2019;49(7):620–7.
Zhang W, et al. KHDC3L mutation causes recurrent pregnancy loss by inducing genomic instability of human early embryonic cells. PLoS Biol. 2019;17(10):e3000468.
Yu Y, et al. Whole-exome sequencing reveals genetic variants in ERC1 and KCNG4 associated with complete hydatidiform mole in Chinese Han women. Oncotarget. 2017;8(43):75264–71.
Kanzi AM, et al. Next generation sequencing and bioinformatics analysis of family genetic inheritance. Front Genet. 2020;11:544162.
Nguyen NMP, et al. The genetics of recurrent hydatidiform moles: new insights and lessons from a comprehensive analysis of 113 patients. Mod Pathol. 2018;31(7):1116–30.
Deveault C, et al. NLRP7 mutations in women with diploid androgenetic and triploid moles: a proposed mechanism for mole formation. Hum Mol Genet. 2009;18(5):888–97.
Wang CM, et al. Identification of 13 novel NLRP7 mutations in 20 families with recurrent hydatidiform mole; missense mutations cluster in the leucine-rich region. J Med Genet. 2009;46(8):569–75.
Begemann M, et al. Maternal variants in NLRP and other maternal effect proteins are associated with multilocus imprinting disturbance in offspring. J Med Genet. 2018;55(7):497–504.
Akoury E, et al. Live births in women with recurrent hydatidiform mole and two NLRP7 mutations. Reprod Biomed Online. 2015;31(1):120–4.
Andreasen L, et al. NLRP7 or KHDC3L genes and the etiology of molar pregnancies and recurrent miscarriage. Mol Hum Reprod. 2013;19(11):773–81.
Messaed C, et al. NLRP7 in the spectrum of reproductive wastage: rare non-synonymous variants confer genetic susceptibility to recurrent reproductive wastage. J Med Genet. 2011;48(8):540–8.
Schilit SLP, et al. SYCP2 translocation-mediated dysregulation and frameshift variants cause human male infertility. Am J Hum Genet. 2020;106(1):41–57.
Yang F, et al. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol. 2006;173(4):497–507.
Nott A, et al. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455(7211):411–5.
Wang W, et al. Overexpression of Uromodulin-like1 accelerates follicle depletion and subsequent ovarian degeneration. Cell Death Dis. 2012;3(11):e433.
Su W, et al. Identification of two mutations in PCDHGA4 and SLFN14 genes in an atrial septal defect family. Curr Med Sci. 2018;38(6):989–96.
Chen P, et al. Differential methylation of genes in individuals exposed to maternal diabetes in utero. Diabetologia. 2017;60(4):645–55.
Acknowledgements
We thank all the patients who participated in this study and the clinicians and embryologists of the Center for Reproductive Medicine, Peking University Third Hospital for supporting.
Funding
This work was supported by the National Natural Science Foundation of China (81971440, 81671458), the National Key Research and Development Program of China (2018YFC1002302), and the Leading Academic Discipline Project of Beijing Education Bureau (BMU20110254).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10815_2022_2592_MOESM1_ESM.xlsx
Supplementary file1 The fertility histories of the 78 unrelatedpatients in our study with at least one HM. (DOCX 31.1 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, J., Yan, L., Li, R. et al. Genetic screening of Chinese patients with hydatidiform mole by whole-exome sequencing and comprehensive analysis. J Assist Reprod Genet 39, 2403–2411 (2022). https://doi.org/10.1007/s10815-022-02592-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-022-02592-z