Abstract
The cryptophyte Rhodomonas sp. is a potential feed source for aquaculture live feed and resource for phycoerythrin (PE) production. This research investigates the influence of light, both quality and quantity, on the biomass productivity, composition and growth rate of Rhodomonas sp. The incident light intensity used in the experiments was 50 μmolphotons m−2 s−1, irrespective of the colour of the light, and cultivation took place in lab-scale flat-panel photobioreactors in turbidostat mode. The highest productivity in volumetric biomass (0.20 gdry weight L−1 day−1), measured under continuous illumination, was observed under green light conditions. Blue and red light illumination resulted in lower productivities, 0.11 gdry weight L−1 day−1 and 0.02 g L−1 day−1 respectively. The differences in production are ascribed to increased absorption of green and blue wavelength by phycoerythrin, chlorophyll and carotenoids, causing higher photosynthetically usable radiation (PUR) from equal photosynthetically absorbed irradiance (PAR). Moreover, phycoerythrin concentration (281.16 mg gDW−1) was stimulated under red light illumination. Because photosystem II (PSII) absorbs poorly red light, the algae had to induce more pigments in order to negate the lower absorption per unit pigment of the incident available photons. The results of this study indicate that green light can be used in the initial growth of Rhodomonas sp. to produce more biomass and, at a later stage, red light could be implemented to stimulate the synthesis of PE. Fourier-transform infrared spectroscopy (FTIR) analysis demonstrated a significant difference between the cells under different light quality, with higher contents of proteins for samples of Rhodomonas sp. cultivated under green light conditions. In comparison, higher carbohydrate contents were observed for cells that were grown under red and blue light.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cryptophyte Rhodomonas is a widely used feed source for many aquaculture species. It has been shown that Rhodomonas contributes to egg production, growth, survival, reproduction and lipid content of copepods, brine shrimps and scallop larvae (Seixas et al. 2009; Guevara et al. 2011; Arndt and Sommer 2014). These copepods, brine shrimps and scallop larvae are the most frequently used and easiest to cultivate feed for fish production in aquaculture. The beneficial aspects of using Rhodomonas as feed are attributed to their favourable content of polyunsaturated fatty acids (PUFAs), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), which are essential for the growth, survival and pigmentation of the aquaculture species mentioned above (Caramujo et al. 2008). The fatty acid content of copepods reflects the fatty acids of the ingested microalgae, hence the importance of the biochemical composition of the microalgal diet (Støttrup et al. 1999).
The ability of Rhodomonas to produce proteins, carotenoids, fatty acids and carbohydrates in a sustainable culturing system makes this alga suitable as feed in the aquaculture sector. In addition, after the extraction of these biological macromolecules, a variety of applications in biomedical, biotechnological and nutraceutical sectors is possible (Manirafasha et al. 2016). The content and type of light-harvesting pigments are essential features of photosynthetic algae species. Rhodomonas contains, besides the common pigments such as chlorophyll a/c and several carotenoids, the water-soluble pigment Cr-phycoerythrin 545 (PE) that harvests light in the green wavelength (λmax = 545 nm) (Doust et al. 2006). The price of PE is highly valuable (30–150 US$ mg−1) depending on the application. Because of its spectral properties (fluorescence emission peak at 578 nm), it is used in its purified form in fluorescence-based detection. In research, some of these applications involve flow cytometry, microarray assays, ELISAs or other applications that require high sensitivity (Chaloub et al. 2015). PE is also used in food colouring and cosmetics (Sudhakar et al. 2015) and linked to anti-parasitic and anti-tumour activities in studies using the marine mollusc Aplysia californica (Coelho et al. 1998). Anti-cancer activities of the three α, β and γ subunits of the PE-containing phycobilisome have also been reported by experiments conducted on mouse tumour cells (Bei et al. 2002).
Optimization of Rhodomonas sp. cultivation has been conducted in aspects of biomass production rate and biochemical composition, but no commercially stable cultivation plan has been applied in aquaculture yet (Seixas et al. 2009; Oostlander et al. 2020; Yamamoto et al. 2020). There is literature focused on the absorption characteristics of PE in Rhodomonas (Van Der Weij-De Wit et al. 2006; da Silva et al. 2009; Marraskuranto et al. 2018), but no literature investigated the growth of this cryptophytic species under different light qualities. This paper fills the gap and provides the knowledge necessary to design a sound cultivation plan for Rhodomonas sp.
Microalgal pigments are dependent on the photon flux density (PFD), the number of photons passing through a particular area per second (μmolphotons m−2 s−1). Consequently, pigmentation depends on the light quality and quantity received during cultivation. During photoacclimation, pigments decrease as a result of increasing light intensities and vice versa (Takache et al. 2012). Moreover, pigment concentration correlates to biomass-specific light absorption rate (Sforza et al. 2015). Strongly absorbed light results in lower pigment concentration.
The goal of our research was to determine the light conditions for optimizing biomass productivity and the PE production for Rhodomonas sp. We investigated optimal light wavelength and intensity for the growth of Rhodomonas sp. Productivity (rx, g L−1 day−1) is defined by the product of the growth rate (μ, day−1) and the biomass concentration (Cx, g L−1). In addition, we investigated how light quality affects PE concentration in Rhodomonas sp. According to the absorption range of the photosynthetic pigment PE (active wavelength 440–570 nm), it is expected that Rhodomonas has the highest production when cultivated using blue/green light (400–600 nm). Similarly, the photopigment composition (biomass composition) would alter with different light quality and quantity. Additionally, weakly absorbed light wavelengths, it is expected to induce the pigment production in Rhodomonas sp. to maximize the light uptake by the antennas of the strain.
Materials and methods
Rhodomonas sp. cultures
Rhodomonas sp. was supplied by the Dutch aquaculture industry, as a strain used in a commercial application. The strain was characterized by 18S sequencing and confirmed as Rhodomonas sp. (Online Resource 1). The marine cryptophytic microalgae Rhodomonas sp. was maintained in pre-sterilized 300-mL Erlenmeyer flasks (20 min at 120 °C) containing 150 mL of 10 times concentrated nutrients of the modified L1 medium to maintain a nutrient-rich condition. The final concentration of the growth medium is 8.82 mM NaNO3, 0.36 mM NaH2PO4·2H2O, 0.11 mM Na2EDTA·2H2O, 0.11 mM FeCl2·6H2O, 9.1 μΜ MnCl2·4H2O, 0.77 μΜ ZnCl2·7H2O, 0.34 μΜ CoCl2·6H2O, 0.4 μΜ CuSO4·5H2O, 0.24 μΜ Na2MoO4·2H2O, 0.29 μΜ vitamin B1, 0.07 μΜ vitamin B12 and 0.01 μΜ vitamin H with a salinity of 30 g L−1 (adapted from Guillard, et al., 1993). The medium was filtered through a Sartorius membrane filter (0.2-μm pore size) into sterilized Erlenmeyer flasks. Cultures were maintained in an orbital incubator at 20 ± 1 °C and 5% CO2 v/vair was supplied in the headspace of the Erlenmeyer flasks. Sodium bicarbonate was added to the L1 medium to a final concentration of 8 mM as a pH buffer. All the stock cultures were continuously illuminated at a photon flux density (PFD) of 120 μmolphotons m−2 s−1 of cool white light provided by TL fluorescent tubes. Culture growth was monitored by measuring the cell abundance with a Coulter counter (Beckman Coulter Z1) to ensure that the inoculum was in the exponential phase before it was used in further experiments.
Experimental conditions
This research consists of two experimental setups. At the first experimental setup, Rhodomonas sp. was cultivated in batch cultures in Erlenmeyer flasks. Different light quantities and qualities were used for this experiment. During the second experimental setup, Rhodomonas sp. was grown in photobioreactors in continuous cultures under a set light intensity with different light wavelengths.
Light quantity
Three experiments were conducted in which Rhodomonas sp. cultures were exposed to different light intensities ranging from 8 ± 10, 60 ± 10 and 80 ± 20 μmolphotons m−2 s−1. Three different wavelengths were applied by white, red, green and blue (WRGB) LED lights, representing the colours blue, green and red (λ peak: 461, 517 and 630 nm respectively) with white LED light (λ range: 415–720 nm) as reference (Fig. 1). The light intensity was measured and monitored manually with the help of a photosynthetically active radiation (PAR) meter (SKP 200/217/140, Skye, UK). Rhodomonas sp. was batch cultured in pre-sterilized 300-mL Erlenmeyer flasks (20 min at 120 °C) containing 150 mL of 10 times concentrated L1 medium (salinity 30 g L−1) and 8 mM sodium bicarbonate at 20 ± 1 °C in an orbital incubator (Gallenkamp) and 5% CO2 V/Vair supply. The experiments were started with a culture density of 4 ± 0.1 × 105 cells mL−1 (10% of medium volume). All experiments were conducted in triplicates. During the experiments, light pollution and background light were excluded.
Light quality
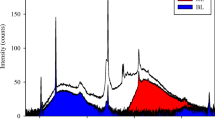
Rhodomonas sp. was continuously cultivated in four different flat panel Algaemist-S photobioreactors (Technical Development Studio, Wageningen University, the Netherlands) with 0.4 L volume, 14 mm light path and 0.028 m2 total illuminated area. The warm light was provided by Bridgelux LED lamps (BXRAW1200, Bridgelux, USA) from one side of the Algaemist-S system. Unintentional exposure to other light sources was prevented by a black cover on the other side of the reactor. In order to illuminate the reactor with different light qualities, the light panel was covered with Lee filters (LEE Filters, USA), allowing transmission of a specific range of wavelengths. The following Lee filters were chosen targeting the absorption peaks of Rhodomonas sp.: Tokyo blue (380–520 nm, λmax peak 445 nm); Aurora Borealis green (520–600 nm, λmax peak 551 nm) and Marius red (λmax peak 700 nm) (Fig. 2). In all cultures, the incident light was 50 μmolphotons m−2 s−1, while in the turbidostat mode, the secondary light PAR sensor of the systems ensured outgoing light of 15 μmolphotons m−2 s−1. After inoculation, the reactor was started running in batch mode until the outgoing light intensity equalled 15–20 μmolphotons m−2 s−1. The temperature within the culture compartment of the photobioreactor (PBR) was kept stable at 22 °C by cooling the adjacent water compartment. The pH was set at 7.5 ± 0.1 and maintained constant by automatically regulating the flow of the air/CO2 mixture pumped into the culture.
Light penetrating the Lee filters, Algaemist LED panel (white) and Rhodomonas sp. absorption spectrum. The corresponding pattern area represents the wavelength range of the light that penetrates through each filter, while the thicker black dashed line represents the absorption spectrum of Rhodomonas sp. The curves illustrate the photon flux density that was employed in the Algaemist experiment, 50 μmolphotons m−2 s−1, when integrated across the entire PAR range (400–700 nm)
Culture analysis
Biomass
Samples were taken daily during light intensity experiments (Erlenmeyers) and wavelength experiments (PBR). These samples were analyzed for basic culture monitoring. Optical density was measured at 750 nm (OD750nm) in a spectrophotometer (HACH, DR 5000), from which biomass concentration (Cx) was calculated as dry weight (Online Resource 2). Cell abundance, size range 7–14 μm, was measured with a Coulter counter (Beckman Coulter Z1). The fact that biomass accumulation was proportional to optical density was shown by a linear relationship between dry weight and OD750nm. All individual sample measurements were performed in triplicate. The growth rate (μ) was calculated for batch cultures from Eq. (1), while for the turbidostat mode as the dilution rate (D) through Eq. (2), where VH is the harvested volume in a period of time and VR the reactor volume.
The biomass production rate (rx) for the turbidostat mode was calculated from the growth rate and the biomass concentration (Cx, Eq. (3)). The biomass production rate and the absorbed light (ΔIph = Iph,in − Iph,out) were used for the calculation of biomass yield on light (Yx/ph) (Eq. (4)). Yx/ph is defined as the biomass concentration that can be produced over a mol of photon.
Phycoerythrin concentration
Samples from each experiment were analyzed on the PE concentration. In the batch culture experiment (Erlenmeyers), PE analysis was performed twice during the growth phase. The first sample was taken when the culture was in the exponential growth phase (day 4), while the second sampling took place during the stationary phase of the culture (day 10). The phycobilin pigments were extracted by a freeze-thawing process, centrifuged and analyzed using UV-VIS spectroscopy according to Bennett and Bogorad (1973) (Lawrenz et al. 2011). Absorbance at 545 nm was used after scatter corrected by subtracting the absorbance at 750 nm. PE was calculated in microgram per litre according to Eq. (5):
where ε is the molar extinction coefficient of PE (2.41 × 106 L mol−1 cm−1), MW is the molecular weight of PE (240,000 g mol−1), d is the path length in centimetre and Vsample and Vbuffer are the volumes of the sample and the buffer respectively. In the PBR experiment, samples for pigment analysis were taken when the culture was in a steady state.
Absorption spectrum
Light absorption was measured in a double beam spectrophotometer (Cary 300 UV-VIS, Agilent, USA) fitted with a Labsphere DRA-CA-3300 integrating sphere. The absolute absorbed light per wavelength was used to calculate the photosynthetically usable radiation (PUR) from PAR using Eq. (6):
where α(λ) represents the probability that a photon with a given wavelength (λ) being absorbed by the cell (Morel 1978). It is derived by the absorption spectrum of Rhodomonas sp., normalized to its peak maximum (λ = 440 nm).
Processing for spectroscopy using FTIR
Fourier-transformed infrared spectrometry (FTIR) analysis was used to investigate changes in the bulk carbohydrate, lipid and protein content. A 50 mL sample was concentrated by centrifuging at 2500 1153×g for 15 min. The supernatant was discarded and 50 mL of ammonium formate (0.5 M) was added to rinse out the salt and preventing osmotic shock. The rinsing was repeated twice. Droplets of the concentrated suspension were placed on a microscope slide and thereafter dried at 50 °C for 24 h. The slides were stored in desiccators until the FTIR analysis, using the PerkinElmer Frontier FTIR equipped with the attenuated total reflectance (ATR) accessory. The spectrum collected was in the range of 4000–650 cm−1, and data was exported using the Unscrambler-X software (Camo Analytics, Norway). The spectral absorption bands were obtained and identified based on previously published studies (Table 1).
Statistical analysis
All data measurements are shown as mean ± standard deviation (± SD) of three independent replicates for the flask experiment, while in the PBR experiments, the measurements that are shown are the average of five daily measurements when the reactor is in a steady state (stable dilution rate). Statistical analysis was performed using SPSS 25.0 statistical package (SPSS Inc., USA) and Prism 8.0.2 (GraphPad, USA). Data were tested for normal distribution (Kolmogorov-Smirnoff goodness of fit test) before being analyzed by ANOVA. The results were analyzed by one-way analyses of variance (ANOVA) with α = 0.05, followed by the post hoc test. For the statistical analysis of the FTIR results, principal component analysis (PCA) was carried out in R, using the factoextra R package to create a ggplot2-based visualization.
Results
Light intensity experiment using batch cultures
Growth
The light intensity of 8 ± 10 μmolphotons m−2 s−1 resulted in low dry weight and cell abundance of Rhodomonas sp. during the cultivation period (Fig. 3a). Cultures incubated under blue light conditions performed better and reached significantly higher biomass concentration on day 10 (p < 0.001), 0.30 ± 0.03 g L−1. In addition, there was no characteristic growth curve (i.e. lag, exponential, stationary, senescent phase) for all the cultures. For 60 ± 10 μmolphotons m−2 s−1 light conditions, the cultures that were exposed to blue light had significantly higher biomass. After 7 days, these cultures had reached the highest density (1.64 ± 0.03 g L−1) compared to the other wavelengths (p = 0.002) (Fig. 3b). In light conditions of 80 ± 20 μmolphotons m−2 s−1, white and blue light resulted in the highest biomass levels, 1.38 ± 0.08 and 1.39 ± 0.04 g L−1 respectively (Fig. 3c). Of all the tested wavelengths of light, blue light led to the highest growth rate in 60 and 80 μmolphotons m−2 s−1 light intensities, 0.63 ± 0.09 day−1. The lowest growth rate in the same light intensities was observed under red light, 0.51 ± 0.02 day−1 (Fig. 4). In 8 μmolphotons m−2 s−1, there was no growth observed for cultures exposed to white and red light, while for blue and green lights, the growth rate was lower than in 60 and 80 μmolphotons m−2 s−1.
Phycoerythrin concentration
Under 8 μmolphotons m−2 s−1 the PE concentration increased significantly between day 4 and day 10 in blue and green wavelength experiments (p = 0.011, p = 0.018). Under blue light the PE content almost doubled from 10 to 19 pg cell−1, whereas under green light, the PE content increased from 9.6 to 16.1 pg cell−1 (Fig. 5a). However, there was no significant change observed in the PE concentration between the exponential and stationary phase in cultures grown under red and white light. Under 60 μmolphotons m−2 s−1 the PE concentration did not follow the same growth pattern as in the cultures under 8 μmolphotons m−2 s−1 (Fig. 5b). During the exponential phase there was no significant difference in the PE content between the cultures grown at different light wavelengths. The concentration did not change in the stationary phase. A 38% reduction of PE concentration in respect to the exponential phase was observed for the cultures that were illuminated by blue light. Under 80 μmolphotons m−2 s−1 all Rhodomonas sp. cultures showed a sharp decrease in PE concentration when they reached the stationary phase, especially for cultures incubated under blue and white light conditions (p = 0.036, p = 0.04) (Fig. 5c).
Phycoerythrin (PE) concentration (average± SD, n = 3, pg cell−1) of Rhodomonas sp. in batch cultures at the exponential (day 4) and stationary phase (day 10), under different light wavelengths (blue, green, red, white) in a low light (8 μmolphotons m−2 s−1), b medium light (60 μmolphotons m−2 s−1) and c high light (80 μmolphotons m−2 s−1) conditions (*indicates the significant difference after ANOVA test and t test of pairwise comparison between the groups ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001)
Light quality experiment
Growth
Continuous Rhodomonas sp. cultures in PBR exposed to different light wavelengths were in a steady state for more than a week. Cultures incubated under green light conditions (peak 550 nm) showed the best absorption by the PE. As a result, volumetric productivity was highest under green light conditions, 0.20 ± 0.05 g L−1 day−1. Cultures exposed to blue and red light, which cannot be well absorbed by PE, showed significant (p < 0.001) lower productivities, 0.11 ± 0.01 and 0.02 ± 0.01 g L−1 day−1, respectively (Fig. 6a). The highest biomass dry weight was reached under white and green light conditions, 0.43 ± 0.05 g L−1 (2.73 106 cells mL−1) and 0.36 ± 0.02 g L−1 (2.67 106 cells mL−1) respectively, significantly higher (p = 0.003) compared to the other wavelengths. Moreover, exposing Rhodomonas sp. to red light resulted in dry weight biomass not higher than 0.04 ± 0.01 g L−1 (0.48 106 cells mL−1), while exposure to blue light resulted in a dry weight of 0.15 ± 0.03 g L−1 (1.11 106 cells mL−1) (Fig. 6b). The growth rate in cultures exposed to blue light was the highest, 0.73 ± 0.11 day−1 (p = 0.004) compared to the rest of the wavelengths (Fig. 6c). The lowest growth rates (p < 0.001) were observed for cultures grown under white light conditions, 0.38 ± 0.04 day−1. The cultures exposed to green and red light conditions demonstrated a slightly higher growth rate, 0.56 ± 0.07 and 0.47 ± 0.14 day−1, respectively. As the PBRs were operated in turbidostat mode, the light absorption was the same for each tested light quality. Therefore, the biomass yield on light followed the same pattern as the volumetric productivity (Fig. 6d). The highest biomass yield on light was calculated for the green light wavelength, 0.91 ± 0.11 g molphotons−1, while the lowest biomass yield on light (p < 0.001) was only 0.13 ± 0.08 g molphotons−1 in red light conditions.
The effect of different wavelengths (blue, green, red, white) on a volumetric productivity rx (g L−1 day−1), b dry weight Cx (g L−1), c growth rate μ (day−1) and d biomass yield on light Yx/ph (g molphotons−1) of Rhodomonas sp. cultivated in PBR reactor (all values average ± SD, n = 7) (*indicates the significant difference after ANOVA test and t test of pairwise comparison with white light as a reference as described in Fig. 5)
Phycoerythrin concentration
When the PBRs were in a steady state samples were taken for the quantification of the PE concentration. The results showed that red light induced the PE accumulation in Rhodomonas sp. (Fig. 7). PE concentration in red light conditions was 281 mg gDW−1 (35.1 pg cell−1), 145% higher than the PE concentrations in cultures exposed to green light conditions, 114 mg gDW−1 (14.3 pg cell−1). The PE concentration measured in cultures grown under white light conditions was the lowest, 23 mg gDW−1 (2.9 pg cell−1). The absorption spectra of Rhodomonas sp. cells in the PBR under different light wavelengths did not present a big difference, except for the red wavelength (Fig. 8).
Phycoerythrin (PE) concentration (average ± SD, n = 3, mg gDW−1) of Rhodomonas sp. cultivated in PBR under different light wavelengths (blue, green, red and white) (*indicates the significant difference after ANOVA test and t test of pairwise comparison with white light as described in Fig. 5)
FTIR analyses
Table 2 demonstrates the band intensities for each treatment, normalized to the highest peak of each spectrum, together with the coefficient of variation (CV). PCA provided information to determine to which extent the different wavelength treatment could be differentiated (Fig. 9). The plot shows a clear separation of the samples of Rhodomonas sp. grown under green and white light conditions, while the samples collected from the incubations under red and blue light are clustered in the same area of PCA. The main variation in PC1 occurs due to the bands that are assigned for proteins and carbohydrates. On the other hand, along PC2, the lipid regions are responsible for the variation. Rhodomonas sp. samples cultivated in green light conditions are separated in the PCA plot, mainly due to the higher concentration of amides. The differentiation between blue and red light incubations is mainly attributed to differences in the carbohydrate content of the samples.
Discussion
Light intensity experiment using batch cultures
Growth
Blue light led to the highest growth rate in 60 and 80 μmolphotons m−2 s−1 light intensities, while the lowest growth rate in the same light intensities was observed under red light. The absence or lower growth in 8 μmolphotons m−2 s−1 confirms results from previous studies (Bartual et al. 2002; Hammer et al. 2002; Chaloub et al. 2015; Vu et al. 2016). The effect of light intensity on the growth of Rhodomonas sp. is explained better by plotting the growth rate against PUR, calculated by all light intensities and wavelengths of the experiment (Fig. 10). The absorption spectrum of Rhodomonas sp. can explain the highest growth rate that was obtained under blue light conditions. The wavelengths of blue light can be absorbed effectively by PE and other pigments. Due to the high absorption of blue light, the PUR in our experiment was higher compared to other light wavelengths. This positive effect of blue light on the growth rate of Rhodomonas sp. is supported by previous research on Rhodomonas salina. Heidenreich and Richardson (2020) reported higher absorption, PUR and growth rates for R. salina under blue light.
The influence of PUR (μmolphotons m−2 s−1) on growth rate μ (average ± SD, n = 3, day−1) of Rhodomonas sp. in batch cultures under different light wavelength (blue, green, red, white). A regression line is fitted through these data points, and the equation describing the regression is presented under the line
A reduction of the biomass of the stationary phase was observed from 60 to 80 μmolphotons m−2 s−1 light conditions in the cultures exposed to blue light, while a biomass increase was noticed for green and red light. Provided that nutrients are not a limiting factor in these high-density cultures, the strongly absorbed blue light is attenuated already in the outermost cells where it could saturate photosynthesis and fails to reach inner cells. In contrast, a more homogenous light climate is obtained for lesser absorbed light qualities, which can be more efficiently used by the cells if it does not saturate photosynthesis. This explanation is supported by Ooms et al. (2017) where they cultivated the cyanobacterium Synechococcus elongatus under different light intensities and wavelengths. They showed that highly absorbed light is not always the best choice for monochromatic cultivation, especially for cultures with high biomass density. Depending on reactor depth, cell density and irradiance, a light wavelength that is not well absorbed, could be the best option to obtain high biomass productivity. This effect can be explained by the fact that weakly absorbed wavelength of light, when biomass concentration is high, can penetrate deeper into the culture and dilute the light better into the reactor.
Phycoerythrin concentration
Pigment adaptation, also called chromatic adaptation in response to light (quantity and quality), is common for macro- and microalgae (Bennett and Bogorad 1973; Schulze et al. 2014) and can be caused by photobleaching (light damage) or physiological response (Gutu and Kehoe 2012). Short-term acclimation (seconds-minutes) involves all kind of reactions to dissipate excess light (Huot and Babin 2010; Hughes et al. 2018), whereas long-term acclimation, the so-called light-shade acclimation, involves changes in the content and ratio of the light-harvesting and protective pigments (Falkowski and Raven 1997). When these processes fail, photobleaching can occur.
A 38% reduction of PE concentration in respect to the exponential phase was observed for the cultures that were illuminated by blue light. This reduction can partly be explained by photoacclimation which occurs in response to PFD, spectral density changes (Falkowski and La Roche 1991) and PFD saturation (Chaloub et al. 2015). As a result, the cultures exposed to the blue light reached the stationary phase the earliest, because of the highest productivity compared to the cultures exposed to the other wavelengths.
In dense cultures, the cells perceive less light due to self-shading, and they become shade adapted. So in dense cultures, which are not limited by nutrients, an increase in pigments is expected to maximize the use of the low light that is available. This hypothesis is in contrast with our observations under 80 μmolphotons m−2 s−1 and can be explained by the fact that a diminution of growth is occurring, probably due to a lack of nutrients, and therefore, less pigmentation is necessary (Lafarga-De la Cruz et al. 2006). Another process should be taken into account when looking at the PE concentration of the cells during the stationary phase of the cultures. The cultures in the present study, both those grown under 60 and 80 μmolphotons m−2 s−1, showed a change in colour from red to yellow/greenish when the stationary and senescence phase was reached. This colour transition is caused by a decrease in pigments and indicates nitrogen starvation (da Silva et al. 2009). The studies of Bartual et al. (2002) and da Silva et al. (2009) show a loss of chlorophylls and PE under nitrogen starvation. These findings support the assumption that nutrients became a limiting factor in our study in the stationary phase for cultures under 60 and 80 μmolphotons m−2 s−1. When nitrogen starvation occurs, PE can function as a source of amino acids for the synthesis of new proteins or protein turnover (Rhiel et al. 1985; da Silva et al. 2009).
The increased PE concentration in Rhodomonas sp. under 8 μmolphotons m−2 s−1 indicates that low light conditions increase the efficiency of cells in an algal culture to capture photons for photosynthesis, which is in agreement with the study of Thinh (1983). As stated by Chaloub et al. (2015), low light intensity triggers algal cells to synthesize more pigments. As a consequence, cultivation with LED lights with low light intensity is considered useful for pigment production for biochemical purposes. da Silva et al. (2009) showed that the chloroplast of the Cryptomonas maculata under low light conditions, around 6 μmolphotons m−2 s−1, contains 50% more of chloroplast area (longitudinal cross-section) than a cell exposed to high light conditions, 20 μmolphotons m−2 s−1. In other words, more pigments are found in a chloroplast under low light conditions. In Rhiel et al. (1985), the morphology of the thylakoids in C. maculata cells, exposed to low and high light (6 μmolphotons m−2 s−1 and 20 μmolphotons m−2 s−1, respectively), was investigated. Their results revealed that 90% of the PE was lost in the cells exposed to high light. The results of our study showed the same phenomenon for Rhodomonas sp., up to 89% of PE is lost when low light is compared to high light (p < 0.001) (Fig. 11).
Phycoerythrin (PE) concentration (average ± SD, n = 3, pg cell−1) of Rhodomonas sp. in batch cultures at the stationary phase (day 10) of experiments under different wavelengths in low light (8 μmolphotons m−2 s−1), medium light (60 μmolphotons m−2 s−1) and high light (80 μmolphotons m−2 s−1) conditions (*indicates the significant difference after t test of pairwise comparison with low light intensity as a reference group as described in Fig. 5)
Light quality experiment
Growth
The incoming PUR of each wavelength treatment can partly explain the growth rate difference. Although in all cultures 50 μmolphotons m−2 s−1 light intensity was applied, the PUR for blue, green, red and white was 45.6, 26.1, 23.6 and 22.1 μmolphotons m−2 s−1 respectively. Even though the total amount of photons absorbed by the reactors was equal for all light qualities except red light, the highly absorbed blue light was absorbed by less cells in the reactors (1.1 106 cells mL−1). As a result, higher light availability per cell could have resulted in a higher growth rate. The positive effect of blue light on the growth rate of R. salina has been reported previously (Heidenreich and Richardson 2020). The growth rates of our study were relatively low, compared to the literature values, where growth rate up to 0.68–0.75 day−1 has been reported for Rhodomonas strains (Lafarga-De la Cruz et al. 2006; Vu et al. 2016). However, these maximum growth rates have been observed in batch cultures during the exponential phase. In our batch experiments, growth rates similar to or higher than the ones reported in the literature were calculated. In the PBR experiments, growth rates were determined by the dilution rate, hence not the maximum but constant growth rates.
Phycoerythrin concentration
Significant lower PE concentrations (p = 0.03) compared to red and green light were measured in the cultures grown under blue light conditions. This effect can be explained from Lafarga-De la Cruz et al. (2006) which states that the reduction of pigment concentration is considered as a process of autoregulation of the photosynthetic apparatus to reach a balance between the gain of light and the demand of energy necessary for microalgae growth. As a result of this phenomenon, the energy in the cultures exposed to blue light was used for growth and not for pigment synthesis.
The absorption spectrum of Rhodomonas sp. cells did not show significant differences when illuminated under different light wavelengths, except for the red wavelength (Fig. 8). According to the PUR calculation, more light should be absorbed in the reactor illuminated by red light than in the reactor illuminated with white light. However, the cell density of the culture in the reactor exposed to red light stayed low, which indicates that the light was not absorbed and the outgoing light was higher compared to the other cultures. This can be described by the spectrum of red light (Fig. 2). Many photons are beyond 680 nm, wavelengths that are not used or used far less efficiently by PSII, because the primary donor of PSII (P680) is not formed (Pavlou et al. 2018). As a result, red light could cause an imbalance in photosynthetic electron transport. It has been reported that R. salina shows inverse chromatic adaptation as a response to the energy imbalance (Lawrenz and Richardson 2017). This strategy induces the production of extra PE and Chl c and decreases the Chl-a content in order to increase the absorption cross-section of PSII and reduces the one of PSI. This explanation is, however, not supported by previous research on energy migration dynamics of Rhodomonas, which suggest that PE545 distributes harvested light equally to PSII and PSI (Van Der Weij-De Wit et al. 2006). We were not able to measure the excitation spectra and determine how the excitation energy of red light is distributed between PSI and PSII.
Conclusions
In an ideal system for high PE production, Rhodomonas sp. should be cultivated with maximum growth rate until the cells become so dense that light becomes limiting. If the culture does not run out of nutrients, the cells will increase their PE concentration. During this research, the highest biomass volumetric productivity and biomass yield on the light of Rhodomonas sp. was obtained in turbidostat mode when it was cultivated under green light conditions. Photopigment composition analyses illustrated that Rhodomonas sp. was rich in PE when it was cultivated under low irradiance or light wavelength that actually was perceived as low irradiance (close to red light). As a result, the green light is recommended for Rhodomonas sp. cultivation as aquaculture feed in order to succeed in a high productivity system and high protein product. On the other hand, when focusing on PE production during the cultivation of Rhodomonas sp., low light intensities and wavelengths that are not well absorbed and have the additional benefit of providing a more homogenous light climate are essential.
Data Availability
Full data are available after a request of the reviewers.
References
Arndt C, Sommer U (2014) Effect of algal species and concentration on development and fatty acid composition of two harpacticoid copepods, Tisbe sp. and Tachidius discipes, and a discussion about their suitability for marine fish larvae. Aquac Nutr 20:44–59
Bartual A, Lubián LM, Gálvez JA, Niell FX (2002) Effect of irradiance on growth, photosynthesis, pigment content and nutrient consumption in dense cultures of Rhodomonas salina (Wislouch) (Cryptophyceae). Ciencias Mar 28:381–392
Bei H, Guang-Ce W, Chen-Kui Z, Zhen-Gang L (2002) The experimental research of R-phycoerythrin subunits on cancer treatment: a new photosensitizer in PDT. Cancer Biother Radiopharm 17:35–42
Bennett A, Bogorad L (1973) Complementary chromatic adaptation in a filamentous blue-green alga. J Cell Biol 58:419–435
Benning LG, Phoenix VR, Yee N, Tobin MJ (2004) Molecular characterization of cyanobacterial silicification using synchrotron infrared micro-spectroscopy. Geochim Cosmochim Acta 68:729–741
Caramujo MJ, Boschker HTS, Admiraal W (2008) Fatty acid profiles of algae mark the development and composition of harpacticoid copepods. Freshw Biol 53:77–90
Chaloub RM, Motta NMS, de Araujo SP, de Aguiar PF, da Silva A (2015) Combined effects of irradiance, temperature and nitrate concentration on phycoerythrin content in the microalga Rhodomonas sp. (Cryptophyceae). Algal Res 8:89–94
Coelho L, Prince JS, Nolen T (1998) Processing of defensive pigment in Aplysia californica: acquisition, modification and mobilization of the red algal pigment, R-phycoerythrin by the digestive gland. J Exp Biol 201:425–42538
da Silva AF, Lourenço SO, Chaloub RM (2009) Effects of nitrogen starvation on the photosynthetic physiology of a tropical marine microalga Rhodomonas sp. (Cryptophyceae). Aquat Bot 91:291–297
Dean AP, Estrada B, Nicholson JM, Sigee DC (2008) Molecular response of Anabaena flos-aquae to differing concentrations of phosphorus: a combined Fourier transform infrared and X-ray microanalytical study. Phycol Res 56:193–201
Doust AB, Wilk KE, Curmi PMG, Scholes GD (2006) The photophysics of cryptophyte light-harvesting. J Photochem Photobiol A 184:1–17
Falkowski PG, La Roche J (1991) Acclimation to spectral irradiance in algae. J Phycol 27:8–14
Falkowski PG, Raven JA (1997) Aquatic photosynthesis. Blackwell Science, Malden, p 375
Giordano M, Heraud P, Beardall J, Wood B, MaNaughton D (2001) Fourier transform infrared spectroscopy as a novel tool to investigate changes in intracellular macromolecular pools in the marine microalga Chaetoceros muellerii (Bacillariophyceae). J Phycol 37:271–279
Guevara M, Bastardo L, Cortez R, Arredondo-Vega B, Romero L, Gomez P (2011) Rhodomonas salina (Cryptophyta) pastes as feed for Brachionus plicatilis (Rotifera). Rev Biol Trop 59:1503–1515
Gutu A, Kehoe DM (2012) Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol Plant 5:1–13
Hammer A, Schumann R, Schubert H (2002) Light and temperature acclimation of Rhodomonas salina (Cryptophyceae): photosynthetic performance. Aquat Microb Ecol 29:287–296
Heidenreich KM, Richardson TL (2020) Photopigment, absorption, and growth responses of marine cryptophytes to varying spectral irradiance. J Phycol 56:507–520
Hughes DJ, Campbell DA, Doblin MA, Kromkamp JC, Lawrenz E, Moore CM, Oxborough K, Prášil O, Ralph PJ, Alvarez MF, Suggett DJ (2018) Roadmaps and detours: active chlorophyll-a assessments of primary productivity across marine and freshwater systems. Environ Sci Technol 52:12039–12054
Huot Y, Babin M (2010) Overview of fluorescence protocols: theory, basic concepts, and practice. In: Suggett DJ, Prasil O, Borowitzka MA (eds) Chlorophyll a fluorescence in aquatic sciences: methods and applications. Springer, Dordrecht, pp 31–74
Lafarga-De la Cruz F, Valenzuela-Espinoza E, Millán-Núñez R, Trees CC, Santamaría-del-Ángel E, Núñez-Cebrero F (2006) Nutrient uptake, chlorophyll a and carbon fixation by Rhodomonas sp. (Cryptophyceae) cultured at different irradiance and nutrient concentrations. Aquac Eng 35:51–60
Lawrenz E, Richardson TL (2017) Differential effects of changes in spectral irradiance on photoacclimation, primary productivity and growth in Rhodomonas salina (Cryptophyceae) and Skeletonema costatum (Bacillariophyceae) in simulated blackwater environments. J Phycol 53:1241–1254
Lawrenz E, Fedewa EJ, Richardson TL (2011) Extraction protocols for the quantification of phycobilins in aqueous phytoplankton extracts. J Appl Phycol 23:865–871
Manirafasha E, Ndikubwimana T, Zeng X, Lu Y, Jing K (2016) Phycobiliprotein: potential microalgae derived pharmaceutical and biological reagent. Biochem Eng J 109:282–296
Marraskuranto E, Raharjo TJ, Kasiamdari RS, Nuringtyas TR (2018) Influence of salinity on growth and phycoerythrin production of Rhodomonas salina. Squalen Bull Mar Fish Postharvest Biotechnol 13:109
Morel A (1978) Available, usable, and stored radiant energy in relation to marine photosynthesis. Deep Sea Res 25:673–688
Ooms MD, Graham PJ, Nguyen B, Sargent EH, Sinton D (2017) Light dilution via wavelength management for efficient high-density photobioreactors. Biotechnol Bioeng 114:1160–1169
Oostlander PC, van Houcke J, Wijffels RH, Barbosa MJ (2020) Optimization of Rhodomonas sp. under continuous cultivation for industrial applications in aquaculture. Algal Res:47. https://doi.org/10.1016/j.algal.2020.101889
Pavlou A, Jacques J, Ahmadova N, Mamedov F, Styring S (2018) The wavelength of the incident light determines the primary charge separation pathway in photosystem II. Sci Rep 8:2837
Rhiel E, Mörschel E, Wehrmeyer W (1985) Correlation of pigment deprivation and ultrastructural organization of thylakoid membranes in Cryptomonas maculata following nutrient deficiency. Protoplasma 129:62–73
Schulze PSC, Barreira LA, Pereira HGC, Perales JA, Varela JCS (2014) Light emitting diodes (LEDs) applied to microalgal production. Trends Biotechnol 32:422–430
Seixas P, Coutinho P, Ferreira M, Otero A (2009) Nutritional value of the cryptophyte Rhodomonas lens for Artemia sp. J Exp Mar Bio Ecol 381:1–9
Sforza E, Calvaruso C, Meneghesso A, Morosinotto T, Bertucco A (2015) Effect of specific light supply rate on photosynthetic efficiency of Nannochloropsis salina in a continuous flat plate photobioreactor. Appl Microbiol Biotechnol 99:8309–8318
Sigee DC, Dean A, Levado E, Tobin MJ (2002) Fourier-transform infrared spectroscopy of Pediastrum duplex: characterization of a micro-population isolated from a eutrophic lake. Eur J Phycol 37:19–26
Støttrup JG, Bell JG, Sargent JR (1999) The fate of lipids during development and cold-storage of eggs in the laboratory-reared calanoid copepod, Acartia tonsa Dana, and in response to different algal diets. Aquaculture 176:257–269
Sudhakar MP, Jagatheesan A, Perumal K, Arunkumar K (2015) Methods of phycobiliprotein extraction from Gracilaria crassa and its applications in food colourants. Algal Res 8:115–120
Takache H, Pruvost J, Cornet JF (2012) Kinetic modeling of the photosynthetic growth of Chlamydomonas reinhardtii in a photobioreactor. Biotechnol Prog 28:681–692
Thinh L-V (1983) Effect of irradiance on the physiology and ultrastructure of the marine cryptomonad, Cryptomonas strain Lis (Cryptophyceae). Phycologia 22:7–11
van der Weij-De Wit CD, Doust AB, van Stokkum IH, Dekker JP, Wilk KE, Curmi PM, Scholes GD, van Grondelle R (2006) How energy funnels from the phycoerythrin antenna complex to photosystem I and photosystem II in cryptophyte Rhodomonas CS24 cells. J Phys Chem B 110:25066–25073
Vu MTT, Douëtte C, Rayner TA, Thoisen C, Nielsen SL, Hansen BW (2016) Optimization of photosynthesis, growth, and biochemical composition of the microalga Rhodomonas salina—an established diet for live feed copepods in aquaculture. J Appl Phycol 28:1485–1500
Yamamoto S, Bossier P, Yoshimatsu T (2020) Biochemical characterization of Rhodomonas sp. Hf-1 strain (cryptophyte) under nitrogen starvation. Aquaculture 516:734648
Funding
This work was financially supported by the ValgOrize project, funded by the INTERREG V 2 Seas Mers Zeeën program, with the financial support of the European Regional Development Fund.
Author information
Authors and Affiliations
Contributions
C. Latsos: conceptualization, methodology, investigation, formal analysis, writing original draft, writing—review and editing. J. van Houcke: conceptualization, writing—review and editing, supervision. L. Blommaert: formal analysis, writing—review and editing. G.P. Verbeeke: methodology, formal analysis, writing—review and editing. J. Kromkamp: writing—review and editing. K.R. Timmermans: conceptualization, writing—review and editing, supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Latsos, C., van Houcke, J., Blommaert, L. et al. Effect of light quality and quantity on productivity and phycoerythrin concentration in the cryptophyte Rhodomonas sp.. J Appl Phycol 33, 729–741 (2021). https://doi.org/10.1007/s10811-020-02338-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-020-02338-3