Abstract
The fluorochrome JC-1 is mainly used in mammalian cells to estimate mitochondrial membrane potential (MMP) as a stress marker, with far less data being available for plants or green algae. To address this, we have validated the possibility for changes in MMP to be used as a sensitive stress indicator in the green alga Chlamydomonas reinhardtii. To optimize the method, we analyzed the conditions for applicable MMP determination, including the proper buffer, excitation/emission wavelengths, and solvent (dimethyl sulfoxide, DMSO) influence on the fluorescence signal, and evaluated usefulness of the method for toxicological research. Our results have allowed us to develop a complete protocol for the estimation of MMP changes in C. reinhardtii. Statistical analyses have confirmed the reproducibility, sensitivity, and applicability of this method for toxicological studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
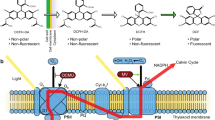
Mitochondria have key roles in energy generation, the metabolism of amino acids and lipids, calcium homeostasis, and cell signaling (Massoz et al. 2017). Many studies have focused on the topic of mitochondrial interactions with other cellular organelles, and their roles in general cell physiology, cell development, programmed cell death, cell pathology, and stress responses (Yao et al. 2004). Mitochondrial membrane potential (MMP) is often studied in living cells using cationic dyes that exhibit fluorescence activity when they accumulate in energized compartments. There are different cell-permeable fluorescent dyes that can measure changes in MMP, such as DiOC6 (3,3′-dihexyloxacarbocyanine iodide), Rh123 (rhodamine-123), tetramethylrhodamine methyl, or JC-1 (5,5,6,6-tetrachloro-1,1,3,3-tetraethylbenzimidazolylcarbocyanine iodide). These dyes are able to accumulate specifically in active mitochondria, in proportion to the magnitude of MMP (Onizuka et al. 2010; Schulze et al. 2010; Shenouda et al. 2010). JC-1 molecules exist in depolarized mitochondria (up to − 100 mV) as monomers that exhibit green fluorescence after excitation at ≈ 490 nm (Reers et al. 1995). At a potential that is more negative than − 140 mV, JC-1 molecules form aggregates (oligomers) that are characterized by orange fluorescence (Reers et al. 1995). This shift in emission wavelengths, due to the formation of dye aggregates, makes the ratio of “orange”-to-“green” fluorescence an excellent parameter for the detection of changes in MMP (Prado et al. 2012).
For many years, the use of JC-1 for flow cytometry, fluorescence microscopy, and fluorescence spectrophotometry has been limited to mammalian cells. Many published protocols examining JC-1 in mammalian cell types, or isolated mitochondria, are currently available (Reers et al. 1995; Cossarizza et al. 1997). Literature reviews also outline the available toxicological studies concerning animal cells, and special attention has been paid to the analysis of MMP changes (Bort et al. 1999; Lim et al. 2006; Kaur and Sanyal 2011; Syed et al. 2016). However, reports on the effects of toxicants on plants are mainly focused on photosynthesis, and they rarely address respiratory processes, or MMP, in particular. Problems with the use of JC-1 in the plant sciences may result from the fact that this dye can be hindered by the plant cell wall, which is a potential barrier for dye molecules, and fluorescence signals can be confounded by chlorophyll, which exhibits autofluorescence. So far, JC-1 staining of plant mitochondria in vivo has only been used in a few basic studies on plant cell bioenergetics, programmed cell death (PCD) investigations, or phytotoxicological experiments (Simeonova et al. 2004; Poborilova et al. 2013; Esperanza et al. 2015). In these studies, different staining protocols and different excitation/emission wavelengths were tested, highlighting the need for this methodology to be optimized for different cell types, experimental conditions, and scientific questions.
The green alga Chlamydomonas reinhardtii is widely used to study the basic fields of cellular biology and molecular biology, such as cell movement, reactions to light, recognition of gametes, cell reactions to biogenic element (carbon, nitrogen, sulfur) concentrations, metabolic pathways, circadian rhythms, or cell cycle progression (Matusiak-Mikulin et al. 2006; Harris 2009). Chlamydomonas reinhardtii is also used in toxicological studies (Aksmann et al. 2014; Majewska et al. 2018; An et al. 2019), because this organism possesses the features of an indicator organism (e.g., high prevalence in the environment, easy cultivation in the laboratory, and high sensitivity to stress factors). Studies on C. reinhardtii cell responses to abiotic stress include classic growth reactions, physiological and biochemical pathways, oxidative stress responses, and changes in the levels of gene transcription and translation (Hema et al. 2007; Aksmann et al. 2014; Majewska et al. 2018). Bearing in mind the usefulness of Chlamydomonas in toxicological studies and the effectiveness of its cell culture model, we have conducted studies to optimize the JC-1 mitochondrial-staining method in alga cells, to make it suitable for the analysis of MMP changes in phytotoxicological studies.
Materials and methods
Materials
Chlamydomonas reinhardtii, strain CC-1690 (wild-type), was purchased from the Chlamydomonas Resource Center (The University of Minnesota, USA). The algae were transferred from slants into a liquid high-salt medium (HSM; pH 6.9 ± 0.1) (Sueoka 1959) and pre-cultured for a few days to adapt them to the conditions of the batch culture. Cultures were grown in 200-mL volume glass test tubes in a water bath at 30 °C, under continuous fluorescent light (250 ± 5 μmol photons m−2 s−1), which was measured inside the test tube (Quantitherm QRT1 light meter, Hansatech Ltd). Cultures with an initial cell density of 1.5 × 106 cells mL−1 were continuously bubbled with sterilized air (PTFE filter, Sartorius 2000) containing 2.5% (v/v) CO2 (Aksmann and Tukaj 2008; Aksmann et al. 2014). Cell number was determined by a particle counter (Beckman Coulter Z2), and cell morphology was observed under a light microscope.
Chemicals and buffers
All chemicals used in the experiments were purchased from Sigma-Aldrich, unless otherwise stated. JC-1 (5,5,6,6-tetrachloro-1,1,3,3-tetraethylbenzimidazolylcarbocyanine iodide) was prepared as a 15 mM stock solution in dimethyl sulfoxide (DMSO) and stored at − 20 °C in small aliquots. The working solution of JC-1 (1.5 mM in DMSO) was freshly prepared before each experiment, except for the JC-1 stability testing, where the working solution was frozen and thawed either once or twice. CCCP (carbonyl cyanide 3-chlorophenylhydrazone; a mitochondrial membrane potential uncoupler) solution (49 mM in DMSO) was stored at − 20 °C in small aliquots and thawed directly before experiments.
PBS buffer (phosphate-buffered saline, consisting of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4; pH 7.4) was chosen based on studies by Prado et al. (2012) and Esperanza et al. (2015). The HEPES (2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid) buffer consisted of 10 mM HEPES, 1 mM CaCl2, 1 mM MgCl2, and 0.6 mM sorbitol (pH 7.2) and was designed to mimic the cytoplasm of C. reinhardtii; this was based on research by Braun and Hegemann (1999). Additives—1 mM CaCl2, 1 mM MgCl2, and 0.6 mM sorbitol, according to Bush and Jones (2008)—were included.
Experimental variants for fluorescence measurements
Aliquots of algal suspension withdrawn from the culture vessel were placed immediately into 15-mL Falcon tubes and spun down (460×g, 5 min). The pellet was flushed twice with an appropriate buffer, and resuspended in the same buffer, to obtain a density of 1.0 × 106 cells mL−1, and used for further analyses. For all fluorescence measurements, a Varioskan Flash microplate reader (Thermo Fisher Scientific, USA) was used.
Comparison of PBS and HEPES buffers
A volume of 0.5 mL alga suspension in either PBS or HEPES buffer was placed in a black Eppendorf tube, and 1 μL of a freshly prepared working solution of JC-1 in DMSO (1.5 mM) was added to obtain a final concentration of 3 μM (Prado et al. 2012). The control sample was an alga suspension with no additives. After a 15 min incubation at 30 °C, a 100 μL volume of cell suspension was placed immediately into a black 96-well plate. Incubation time was based on prior literature (Prado et al. 2012), while temperature was chosen based on algal culture conditions. The fluorescence was measured within the following wavelengths: excitation wavelength (λex) = 420–500 nm and emission wavelength (λem) = 525 nm and 600 nm for monomers and oligomers, respectively (Prado et al. 2012).
Characteristics of the excitation/emission fluorescence spectra
An alga suspension in HEPES buffer, a control, and a sample with JC-1 in DMSO were prepared and incubated, as described in “Comparison of PBS and HEPES buffers.” The fluorescence was measured at (1) λex = 470–510 nm, λem = 538, 544, and 596 nm; (2) λex = 488, 500, and 508 nm, λem = 530–610 nm.
The influences of CCCP and DMSO on the fluorescence signal
An alga suspension in HEPES buffer, the control, and those with JC-1 in DMSO were prepared and incubated, as described in “Comparison of PBS and HEPES buffers.” To create a strong, single-positive green fluorescence control, CCCP-containing samples were prepared according to Prado et al. (2012). In brief, 0.5 μL of a 49 mM solution of CCCP in DMSO was added to 0.5 mL of JC-1 alga suspension, to obtain a final CCCP concentration of 49 μM. This was much the same method by which the influence of DMSO on the fluorescence signal was determined; 0.5 μL of DMSO sample was added instead of the sample of CCCP in DMSO. The fluorescence was measured at λex = 488 nm and λem = 538 nm and 596 nm for monomers and oligomers, respectively.
JC-1 working solution stability
The alga suspensions in HEPES buffer—the control suspension and the sample with JC-1 in DMSO—were prepared and incubated as described in “Comparison of PBS and HEPES buffers.” Fluorescence was measured at λex = 488 nm and λem = 530–615 nm. The JC-1 working solution was then frozen, stored at − 20 °C, and subjected to two cycles of freezing/thawing. After each cycle the experiment was repeated.
The usefulness of the method for phytotoxicity testing
To confirm the usefulness of the method for toxicological analyses, we compared the control cells with the cells treated with diclofenac (DF) (2-[2-(2,6-dichloroanilino)phenyl]acetic acid) using a protocol that was based on the results of the above experiments. For this purpose, algae were cultured as described in “Materials” above. For experimental variants that were designed for toxicity testing, the algae were exposed for 3 h to different concentrations of DF, corresponding to the toxicological values EC25 (221.4 μM), EC50 (425.9 μM), and EC75 (686.4 μM). These concentrations were calculated on the basis of the inhibition of C. reinhardtii population growth in our earlier work (Majewska et al. 2018). After this, cell suspension samples were taken from both the control and the DF-treated cultures, and mitochondrial membrane potential was examined, as described above. For an illustration of the method’s effect, pictures of the C. reinhardtii control and the DF-treated cells were also prepared, using epifluorescence microscopy. Orange/green fluorescence was measured, as an indicator of the depolarization of the mitochondrial membrane.
Statistics
Statistical analyses were performed using OriginPro 2018 (OriginLab Corporation) and Statistica 9.0 (StatSoft). All of the data were expressed as means of at least three independent experiments ± SE or SD. Statistica 9.0 (StatSoft) was used for confirming the representativeness of the obtained results, as well as for canonical discriminate analysis, used to plot a graph of individuals on the discriminate dimensions. The same software was applied to compute a basic statistic, Student’s t test for normal distribution, and the non-parametric Mann–Whitney test for analytical groups, when the population could not be assumed to be normally distributed. A p value < 0.05 was considered to be significant.
Results and discussion
Data from the literature indicate that JC-1 has been successfully used to estimate MMP in animal cells (Cossarizza et al. 1997; Holtsberg et al. 1998) with much less data being available for plants or green algae (Simeonova et al. 2004; Esperanza et al. 2015). In contrast to animal cells, plant cells and microalgae are surrounded by cell walls, which are complex structures that can affect the efficiency of organelle staining. Considering that MMP in living cells is very sensitive to stress factors, we decided to validate the possibility of MMP use as a sensitive indicator of changes in the physiological status of C. reinhardtii. To optimize the method for this purpose, we chose the optimal conditions for applicable MMP determination (i.e., the proper buffer, the correct excitation/emission wavelengths, JC-1 working solution stability, and the influence of DMSO on the fluorescence signal).
In papers describing the use of JC-1 for MMP determination, both the HEPES and PBS buffers are recommended (Simeonova et al. 2004; Esperanza et al. 2015). We have found that the JC-1 fluorescence intensity differs significantly, depending on the buffer that is used for alga incubation (Fig. 1). JC-1 oligomer fluorescence in the HEPES buffer appeared to be more than three times higher than that in PBS. These changes are in line with the work by Tomasek et al. (2018), which determined that PBS, as a flow cytometry buffer, provokes an increase in Escherichia coli cell membrane permeability, causing a fluorescence signal loss that is related to the viability and physiological activity of the bacteria (Tomasek et al. 2018). Thus, we can assume that the increased membrane permeability caused by PBS results in better permeability for the JC-1 monomers, but it also causes an inhibition of respiratory activity in the C. reinhardtii cells. As a result, the oligomer-derived fluorescence signals in the PBS buffer were relatively low compared with those in HEPES, and so we chose HEPES for all further experiments.
In the literature, different excitation/emission maxima for the JC-1 measurements are given: usually 488 nm for excitation and between 525 and 600 nm for emission (Prado et al. 2012; Esperanza et al. 2015). Thus, we decided to scan the JC-1 fluorescence signal in vivo to find the best excitation and emission wavelengths for our experiments (Fig. 2). For the estimation of MMP, because the ratio of JC-1 oligomers (orange fluorescence) to monomers (green fluorescence) needs to be calculated (Esperanza et al. 2015), we analyzed different excitation/emission wavelength combinations to obtain the highest value of the ratio. Although an excitation wavelength of 508 nm gave the highest value of fluorescence (Fig. 2b), the following combination was chosen: λex = 488 nm and λem = 538 nm (green fluorescence, monomers) or λem = 596 nm (orange fluorescence, oligomers), because it resulted in the highest orange/green fluorescence ratios (Fig. 2a).
To control the specificity of JC-1 staining, we applied CCCP, a proton ionophore that eliminates the mitochondrial transmembrane potential. A reduction in the ratio of orange-to-green fluorescence intensities (Table 1) in CCCP-treated cells indicates the inhibition of JC-1 aggregate formation, due to a decrease of MMP, and confirms that JC-1 does not bind to cellular components other than the mitochondria (Simeonova et al. 2004). This interpretation was confirmed by the use of canonical discrimination analysis (Fig. 3), which showed that all three analytical groups, namely non-stained cells, JC-1-stained cells, and JC-1 + CCCP–treated cells, were clearly distinguished. Thus, we can conclude that the method is repeatable and well-adapted to measuring the target of interest (Fig. 3).
Because DMSO was used as a solvent for JC-1 and CCCP, an experiment was performed to estimate the possible influence of DMSO itself on the florescence signal. We found no statistically significant differences between the group with JC-1 and the group with an additional portion (0.5 μL) of DMSO. However, the variation of these results was lower in the group without additional DMSO (Table 1). This indicates that the final DMSO concentration of 0.3% that was used in the JC-1 samples was not high enough to have any effects on the experiment, and thus, this fraction should not be increased any further. The high degree of error that was observed in the samples with additional DMSO could be the result of an increase in the membrane permeability for JC-1, which would make staining less selective and less reproducible.
In the next step of the experiment, JC-1 working solution stability during freezing/thawing cycle was examined. It was found that after one freezing/thawing cycle, followed by storage at − 20 °C, the JC-1 working solution efficiency was significantly decreased, and the fluorescence intensity was reduced by almost twofold (Fig. 4). Also, the fluorescence intensity index (596/538 nm) in the control C. reinhardtii cells changed from 28.35 ± 5.6 for fresh JC-1 working solution to 17.67 ± 0.3 after one cycle of freezing/thawing, and then to 13.71 ± 0.6 after two cycles of freezing/thawing of working solution. The manufacturers’ protocols for JC-1 staining recommend against freezing the JC-1 working solution (it is recommended to store stock solution in small aliquots that should be used only once), but the degree of reactivity loss from freezing is not given (Abcam 2017). Our experiments indicate that the diminishment in the fluorescence signal caused by freezing/thawing of JC-1 working solution is large enough to render the measurements unreliable and that this dye should only be stored as aliquots of stock solution thawed immediately before use.
The in vivo fluorescence emission spectra of fresh JC-1 working solution and once frozen/thawed JC-1 working solution. Algae were incubated in HEPES buffer, excitation λ = 488 nm, emission λ = 530–615 nm. Error bar values (or ±) indicate SD, n = 9. The fluorescence intensity index for fresh JC-1 working solution and for once and twice frozen/thawed JC-1 working solution is shown below the graph
To confirm the usefulness of the JC-1 method for phytotoxicity testing, we used DF, a non-steroidal anti-inflammatory drug that is found as a contaminant in water reservoirs worldwide (Trombini et al. 2019; Zaborska et al. 2019). This substance has been found to be toxic for plants and algae (Ferrari et al. 2003; Majewska et al. 2018). Here, to link the assumed MMP changes with the population growth inhibition caused by DF, we chose the DF concentrations that correspond to toxicological values EC25, EC50, and EC75, which were found in our previous work to inhibit the growth of C. reinhardtii population by 25, 50, and 75%, respectively (Majewska et al. 2018). The results indicate that our protocol enables observations of MMP interruptions, as caused by DF in C. reinhardtii cells, and that MMP is sensitive to changes in DF concentration (Fig. 5). In cells treated by DF at EC25 concentration, MMP decreased by about 30%. In EC50- and EC75-treated cells, MMP was diminished by about 60 and 80%, respectively (Fig. 5a). These results suggest that one of the important factors in alga growth inhibition caused by DF is respiration disorder. This may result from the possible actions of this drug as a calcium homeostasis disruptor, analogous to DF action in animal cells (Gan 2010). Moreover, the results indicate that MMP measurements can be a rapid and sensitive indicator of DF phytotoxicity.
a The fluorescence intensity index (596/538 nm) for the four analytical groups of Chlamydomonas reinhardtii cells: those treated with DF at concentrations corresponding to the toxicological values of EC25, EC50, and EC75, and a control group. Asterisk indicates statistically significant differences compared with the control group at p < 0.05, the Mann−Whitney test. b JC-1 fluorescence in the cells, visualized using epifluorescence microscopy. The green and orange fluorescence signals are merged
Based on our results, we were able to develop a complete protocol for the estimation of MMP changes in C. reinhardtii cells (see “The MMP assay protocol for Chlamydomonas reinhardtii cells, using JC-1”). Our statistical analyses confirm that the results are reproducible and that the method is sensitive and suitable for toxicological studies.
The MMP assay protocol for Chlamydomonas reinhardtii cells, using JC-1
-
1.
Centrifuge the Chlamydomonas reinhardtii cell suspension (1.0 × 106 cells mL−1) at 460×g for 5 min.
-
2.
Wash the pellet gently in HEPES buffer: 10 mM HEPES, 1 mM CaCl2, 1 mM MgCl2, 0.6 mM sorbitol, pH 7.2; and centrifuge again under the same conditions.
-
3.
Resuspend the cells gently in HEPES buffer (1.0 × 106 cells mL−1).
-
4.
Prepare fresh JC-1 working solution (1.5 mM in DMSO) and the CCCP solution (49 mM in DMSO).
-
5.
Place 500 μL of the alga suspension in the HEPES buffer, in black Eppendorf tubes, and add 0.5 μL of CCCP solution (49 mM). Mix gently and incubate for 15 min at 30 °C. Add 1 μL of JC-1 working solution (1.5 mM). Mix gently and incubate for 15 min at 30 °C.
-
6.
For fluorescence measurements, pipette 100 μL of each sample into a black 96-well plate.
-
7.
Measure the fluorescence at λex = 488 nm and at λem = 538 nm (monomers) or 596 nm (oligomers), using a microplate reader.
-
8.
Determine the ratio between the JC-1 oligomers and monomers.
References
Abcam (2017) JC1 -mitochondrial membrane potential assay kit (ab113850 ) version 3. www.abcam.com
Aksmann A, Tukaj Z (2008) Intact anthracene inhibits photosynthesis in algal cells: a fluorescence induction study on Chlamydomonas reinhardtii cw92 strain. Chemosphere 74:26–32
Aksmann A, Pokora W, Baścik-Remisiewicz A, Dettlaff-Pokora A, Wielgomas B, Dziadziuszko M, Tukaj Z (2014) Time-dependent changes in antioxidative enzyme expression and photosynthetic activity of Chlamydomonas reinhardtii cells under acute exposure to cadmium and anthracene. Ecotoxicol Environ Saf 110:31–40
An C, Huang G, Weger H, Raina-Fulton R, Weger H (2019) Insights into long-term toxicity of triclosan to freshwater green algae in Lake Erie. Environ Sci Technol 53:2189–2198
Bort R, Ponsoda X, Jover R, Gómez-Lechón MJ, Castell JV (1999) Diclofenac toxicity to hepatocytes: a role for drug metabolism in cell toxicity. J Pharmacol Exp Ther 288:65–72
Braun FJ, Hegemann P (1999) Direct measurement of cytosolic calcium and pH in living Chlamydomonas reinhardtii cells. Eur J Cell Biol 78:199–208
Bush DS, Jones RL (2008) Measuring intracellular Ca2+ levels in plant cells using the fluorescent probes, indo-1 and fura-2: progress and prospects. Plant Physiol 93:841–845
Cossarizza A, Ortolani C, Monti D, Franceschi C (1997) Cytometric analysis of immunosenescence. Cytometry 27:297–313
Esperanza M, Cid Á, Herrero C, Rioboo C (2015) Acute effects of a prooxidant herbicide on the microalga Chlamydomonas reinhardtii: screening cytotoxicity and genotoxicity endpoints. Aquat Toxicol 165:210–221
Ferrari B, Paxéus N, Giudice RL, Pollio A, Garric J (2003) Ecotoxicological impact of pharmaceuticals found in treated wastewaters: study of carbamazepine, clofibric acid, and diclofenac. Ecotoxicol Environ Saf 55:359–370
Gan TJ (2010) Diclofenac: an update on its mechanism of action and safety profile. Curr Med Res Opin 26:1715–1731
Harris EH (2009) The Chlamydomonas sourcebook: introduction to Chlamydomonas and its laboratory use. Academic Press, New York
Hema R, Senthil-Kumar M, Shivakumar S, Chandrasekhara Reddy P, Udayakumar M (2007) Chlamydomonas reinhardtii, a model system for functional validation of abiotic stress responsive genes. Planta 226:655–670
Holtsberg F, Steiner M, Keller J et al (1998) Lysophosphatidic acid induces necrosis and apoptosis in hippocampal neurons. J Neurochem 70:66–76
Kaur J, Sanyal SN (2011) Intracellular pH and calcium signaling as molecular targets of diclofenac-induced apoptosis against colon cancer. Eur J Cancer Prev 20:263–276
Lim MS, Lim PLK, Gupta R, Boelsterli UA (2006) Critical role of free cytosolic calcium, but not uncoupling, in mitochondrial permeability transition and cell death induced by diclofenac oxidative metabolites in immortalized human hepatocytes. Toxicol Appl Pharmacol 217:322–331
Majewska M, Harshkova D, Guściora M, Aksmann A (2018) Phytotoxic activity of diclofenac: evaluation using a model green alga Chlamydomonas reinhardtii with atrazine as a reference substance. Chemosphere 209:989–997
Massoz S, Hanikenne M, Bailleul B, Coosemans N, Radoux M, Miranda-Astudillo H, Cardol P, Larosa V, Remacle C (2017) In vivo chlorophyll fluorescence screening allows the isolation of a Chlamydomonas mutant defective for NDUFAF3, an assembly factor involved in mitochondrial complex I assembly. Plant J 92:584–595
Matusiak-Mikulin K, Tukaj C, Tukaj Z (2006) Relationships between growth, development and photosynthetic activity during the cell cycle of Desmodesmus armatus (Chlorophyta) in synchronous cultures. Eur J Phycol 41:29–38
Onizuka S, Tamura R, Hosokawa N, Kawasaki Y, Tsuneyoshi I (2010) Local anesthetics depolarize mitochondrial membrane potential by intracellular alkalization in rat dorsal root ganglion neurons. Anesth Analg 111:775–783
Poborilova Z, Opatrilova R, Babula P (2013) Toxicity of aluminium oxide nanoparticles demonstrated using a BY-2 plant cell suspension culture model. Environ Exp Bot 91:1–11
Prado R, Rioboo C, Herrero C, Cid A (2012) Screening acute cytotoxicity biomarkers using a microalga as test organism. Ecotoxicol Environ Saf 86:219–226
Reers M, Smiley ST, Mottola-Hartshorn C, Chen A, Lin M, Chen LB (1995) Mitochondrial membrane potential monitored by JC-1 dye. Methods Enzymol 260:406–414
Schulze T, Weiss S, von der Schymanski E, Ohe PC, Schmitt-Jansen M, Altenburger R, Streck G, Brack W (2010) Identification of a phytotoxic photo-transformation product of diclofenac using effect-directed analysis. Environ Pollut 158:1461–1466
Shenouda SM, Weihrauch D, Kluge MA, Hagen TM, Smith AR, Kizhakekuttu TJ, Kluge MA, Weihrauch D, Gutterman DD, Vita JA (2010) Altered mitochondrial membrane potential, mass, and morphology in the mononuclear cells of humans with type 2 diabetes. Transl Res 156:15–25
Simeonova E, Garstka M, Kozioł-Lipińska J, Mostowska A (2004) Monitoring the mitochondrial transmembrane potential with the JC-1 fluorochrome in programmed cell death during mesophyll leaf senescence. Protoplasma 223:143–153
Sueoka N (1959) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci 46:83–91
Syed M, Skonberg C, Hansen SH (2016) Mitochondrial toxicity of diclofenac and its metabolites via inhibition of oxidative phosphorylation (ATP synthesis) in rat liver mitochondria: possible role in drug induced liver injury (DILI). Toxicol in Vitro 31:93–102
Tomasek K, Bergmiller T, Guet CC (2018) Lack of cations in flow cytometry buffers affect fluorescence signals by reducing membrane stability and viability of Escherichia coli strains. J Biotechnol 268:40–52
Trombini C, Hampel M, Blasco J (2019) Assessing the effect of human pharmaceuticals (carbamazepine, diclofenac and ibuprofen) on the marine clam Ruditapes philippinarum: An integrative and multibiomarker approach. Aquat Toxicol 208:146–156
Yao N, Eisfelder BJ, Marvin J, Greenberg JT (2004) The mitochondrion - An organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J 40:596–610
Zaborska A, Siedlewicz G, Szymczycha B, Dzierzbicka-Głowacka L, Pazdro K (2019) Legacy and emerging pollutants in the Gulf of Gdańsk (southern Baltic Sea) – loads and distribution revisited. Mar Pollut Bull 139:238–255
Acknowledgments
We would like to thank Dr. Małgorzata Kapusta (Department of Plant Cytology and Embryology, Faculty of Biology, University of Gdansk) for providing the fluorescence microscopy photographs of the Chlamydomonas cells.
Funding
This work was supported by the National Science Center, Poland [grant numbers UMO-2016/23/B/NZ9/00963].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Harshkova, D., Zielińska, E. & Aksmann, A. Optimization of a microplate reader method for the analysis of changes in mitochondrial membrane potential in Chlamydomonas reinhardtii cells using the fluorochrome JC-1. J Appl Phycol 31, 3691–3697 (2019). https://doi.org/10.1007/s10811-019-01860-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-019-01860-3