Abstract
Purpose
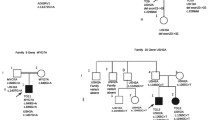
This study investigated the new splice site mutations of Myosin VIIA (MYO7A) in patients with Usher syndrome type 1 (USH1) from a three-generation Chinese consanguineous family.
Methods
All subjects underwent comprehensive ophthalmic examinations and an audiometric test. Demographic data, family history, and peripheral blood leukocytes were collected. We performed whole exome sequencing (WES) to analyze the genomic DNA of the family. DNA sequence and restriction fragment length polymorphism (RFLP) analyses were also done. The identified genetic variants were validated by conducting polymerase chain reaction (PCR) in 100 healthy control subjects and comparing with the NCBI VARIANT database and the 1000 Genomes Project. The functional consequences were further analyzed.
Results
WES identified two new splice site mutations (c.5648G > A(rs111033215) and c.6238-1G > C) in MYO7A in two patients with USH1, i.e., the proband and her elder brother. DNA sequence and RFLP analyses showed that other members without USH1 carried only one of the two mutations. In the analysis of healthy controls, neither mutation existed. Both mutations were predicted to be damaging and were most likely associated with USH1.
Conclusion
In the three-generation Chinese consanguineous family with USH1, c.5648G > A(rs111033215) and c.6238-1G > C mutations in MYO7A are most likely associated with the disease. Our findings expand the mutational spectrum of MYO7A, which will enhance the understanding of the genetic abnormalities in USH1 and provide more evidence for future investigations on therapeutic strategies such as precise gene replacement or gene editing.





Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available in the NCBI ClinVar repository, ClinVar accession number: SCV002510626, SCV002508889.
References
Toms M, Pagarkar W, Moosajee M (2020) Usher syndrome: clinical features, molecular genetics and advancing therapeutics. Ther Adv Ophthalmol 12:2515841420952194. https://doi.org/10.1177/2515841420952194
Mathur P, Yang J (2015) Usher syndrome: hearing loss, retinal degeneration and associated abnormalities. Biochim Biophys Acta 1852:406–420. https://doi.org/10.1016/j.bbadis.2014.11.020
Kimberling WJ, Hildebrand MS, Shearer AE, Jensen ML, Halder JA, Trzupek K, Cohn ES, Weleber RG, Stone EM, Smith RJ (2010) Frequency of Usher syndrome in two pediatric populations: implications for genetic screening of deaf and hard of hearing children. Genet Med 12:512–516. https://doi.org/10.1097/GIM.0b013e3181e5afb8
Jouret G, Poirsier C, Spodenkiewicz M, Jaquin C, Gouy E, Arndt C, Labrousse M, Gaillard D, Doco-Fenzy M, Lebre AS (2019) Genetics of Usher Syndrome: new insights from a meta-analysis. Otol Neurotol 40:121–129. https://doi.org/10.1097/mao.0000000000002054
Reiners J, Nagel-Wolfrum K, Jürgens K, Märker T, Wolfrum U (2006) Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res 83:97–119. https://doi.org/10.1016/j.exer.2005.11.010
Jatana KR, Thomas D, Weber L, Mets MB, Silverman JB, Young NM (2013) Usher syndrome: characteristics and outcomes of pediatric cochlear implant recipients. Otol Neurotol 34:484–489. https://doi.org/10.1097/MAO.0b013e3182877ef2
Géléoc GGS, El-Amraoui A (2020) Disease mechanisms and gene therapy for Usher syndrome. Hear Res 394:107932. https://doi.org/10.1016/j.heares.2020.107932
Dad S, Rendtorff ND, Tranebjærg L et al (2016) Usher syndrome in Denmark: mutation spectrum and some clinical observations. Mol Genet Genom Med 4:527–539. https://doi.org/10.1002/mgg3.228
Eisenberger T, Slim R, Mansour A et al (2012) Targeted next-generation sequencing identifies a homozygous nonsense mutation in ABHD12, the gene underlying PHARC, in a family clinically diagnosed with Usher syndrome type 3. Orphanet J Rare Dis 7:59. https://doi.org/10.1186/1750-1172-7-59
Jian X, Boerwinkle E, Liu X (2014) In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res 42:13534–13544. https://doi.org/10.1093/nar/gku1206
Liu X, Wu C, Li C, Boerwinkle E (2016) dbNSFP v3.0: a One-Stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 37:235–241. https://doi.org/10.1002/humu.22932
Ji RX, Su J, Guo XS, Yu ZQ, Liu HX, Liu CQ (2013) Clinical study on the effect of alendronate sodium in the prevention of second fracture in patients with osteoporotic vertebral fracture. Zhejiang J Trauma Surg 18:823–825
Ouyang XM, Yan D, Du LL et al (2005) Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum Genet 116:292–299. https://doi.org/10.1007/s00439-004-1227-2
Cremers FP, Kimberling WJ, Külm M et al (2007) Development of a genotyping microarray for Usher syndrome. J Med Genet 44:153–160. https://doi.org/10.1136/jmg.2006.044784
Nakanishi H, Ohtsubo M, Iwasaki S, Hotta Y, Takizawa Y, Hosono K, Mizuta K, Mineta H, Minoshima S (2010) Mutation analysis of the MYO7A and CDH23 genes in Japanese patients with Usher syndrome type 1. J Hum Genet 55:796–800. https://doi.org/10.1038/jhg.2010.115
Bonnet C, Grati M, Marlin S et al (2011) Complete exon sequencing of all known Usher syndrome genes greatly improves molecular diagnosis. Orphanet J Rare Dis 6:21. https://doi.org/10.1186/1750-1172-6-21
Jacobson SG, Cideciyan AV, Gibbs D et al (2011) Retinal disease course in Usher syndrome 1B due to MYO7A mutations. Investig Ophthalmol Vis Sci 52:7924–7936. https://doi.org/10.1167/iovs.11-8313
Le QuesneStabej P, Saihan Z, Rangesh N et al (2012) Comprehensive sequence analysis of nine Usher syndrome genes in the UK National Collaborative Usher Study. J Med Genet 49:27–36. https://doi.org/10.1136/jmedgenet-2011-100468
Bonnet C, Riahi Z, Chantot-Bastaraud S et al (2016) An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients. Eur J Hum Genet 24:1730–1738. https://doi.org/10.1038/ejhg.2016.99
Fokkema I, Kroon M, López Hernández JA, Asscheman D, Lugtenburg I, Hoogenboom J, den Dunnen JT (2021) The LOVD3 platform: efficient genome-wide sharing of genetic variants. Eur J Hum Genet 29:1796–1803. https://doi.org/10.1038/s41431-021-00959-x
Valero R, de Castro-Miró M, Jiménez-Ochoa S, Rodríguez-Ezcurra JJ, Marfany G, Gonzàlez-Duarte R (2019) Aberrant splicing events associated to CDH23 noncanonical splice site mutations in a proband with atypical Usher syndrome 1. Genes (Basel). https://doi.org/10.3390/genes10100732
Cheng L, Yu H, Jiang Y, He J, Pu S, Li X, Zhang L (2018) Identification of a novel MYO7A mutation in Usher syndrome type 1. Oncotarget 9:2295–2303. https://doi.org/10.18632/oncotarget.23408
Petit C (2001) Usher syndrome: from genetics to pathogenesis. Annu Rev Genom Hum Genet 2:271–297. https://doi.org/10.1146/annurev.genom.2.1.271
Boëda B, El-Amraoui A, Bahloul A et al (2002) Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. Embo J 21:6689–6699. https://doi.org/10.1093/emboj/cdf689
Lopes VS, Gibbs D, Libby RT, Aleman TS, Welch DL, Lillo C, Jacobson SG, Radu RA, Steel KP, Williams DS (2011) The Usher 1B protein, MYO7A, is required for normal localization and function of the visual retinoid cycle enzyme, RPE65. Hum Mol Genet 20:2560–2570. https://doi.org/10.1093/hmg/ddr155
Calabro KR, Boye SL, Choudhury S et al (2019) A novel mouse model of MYO7A USH1B reveals auditory and visual system haploinsufficiencies. Front Neurosci 13:1255. https://doi.org/10.3389/fnins.2019.01255
Acknowledgements
We thank Medjaden Inc for scientific editing of this manuscript.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
QL and XZ: data curation; formal analysis; writing—original draft. DY: investigation; methodology; project administration. ZS: Software. XZ: Writing—review and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
This study was approved by the institutional review board of the Eye and ENT Hospital of Fudan University (Shanghai, China), and was conducted in compliance with the Declaration of Helsinki.
Informed consent
Written informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, Q., Yang, D., Shen, Z. et al. New splice site mutations in MYO7A causing Usher syndrome type 1: a study on a Chinese consanguineous family. Int Ophthalmol 43, 2091–2099 (2023). https://doi.org/10.1007/s10792-022-02611-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10792-022-02611-z