Abstract
Platyrrhine (New World) monkeys possess highly polymorphic color vision owing to allelic variation of the single-locus L/M opsin gene on the X chromosome. Most species consist of female trichromats and female and male dichromats. Howlers (genus Alouatta) are an exception; they are considered to be routinely trichromatic with L and M opsin genes juxtaposed on the X chromosome, as seen in catarrhine primates (Old World monkeys, apes, and humans). Yet it is not known whether trichromacy is invariable in howlers. We examined L/M opsin variation in wild howler populations in Costa Rica and Nicaragua (Alouatta palliata) and Belize (A. pigra), using fecal DNA. We surveyed exon 5 sequences (containing the diagnostic 277th and 285th residues for λmax) for 8 and 18 X chromosomes from Alouatta palliata and A. pigra, respectively. The wavelengths of maximal absorption (λmax) of the reconstituted L and M opsin photopigments were 564 nm and 532 nm, respectively, in both species. We found one M–L hybrid sequence with a recombinant 277/285 haplotype in Alouatta palliata and two L–M hybrid sequences in A. pigra. The λmax values of the reconstituted hybrid photopigments were in the range of 546~554 nm, which should result in trichromat phenotypes comparable to those found in other New World monkey species. Our finding of color vision variation due to high frequencies of L/M hybrid opsin genes in howlers challenges the current view that howlers are routine and uniform trichromats. These results deepen our understanding of the evolutionary significance of color vision polymorphisms and routine trichromacy and emphasize the need for further assessment of opsin gene variation as well as behavioral differences among subtypes of trichromacy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primates are unique among the placental mammals in having trichromatic color vision (Jacobs 2008). This is achieved by the presence of two subtypes of the X-chromosomal long to middle wavelength-sensitive (L/M) opsin genes in addition to an autosomal short wavelength sensitive (S) opsin gene. In catarrhine primates (Old World monkeys, apes, and humans), the two subtypes, L and M opsin genes, are juxtaposed on the X chromosome. Therefore, not only females but also males with a single X chromosome have both L and M opsin genes and are thus capable of trichromatic color vision. However, in most species of platyrrhine primates (New World monkeys), subtypes of the L/M opsin are provided by allelic variation of a single-locus L/M opsin gene on the X chromosome, rendering females either trichromatic or dichromatic and males dichromatic.
Howlers (Alouatta) are an exception among New World monkeys in having both an L and an M opsin gene on the same X chromosome (Jacobs et al. 1996). This gene rearrangement is thought to have occurred independently from the similar gene rearrangement in catarrhine primates because of distinct localization patterns of a cis regulatory region, known as the locus control region (LCR) (Dulai et al. 1999). In the case of catarrhine L/M opsin genes, a single LCR is situated upstream of the L/M opsin gene array and controls expression of both L and M opsin genes, whereas in howlers the LCR is situated upstream of each of the L and M opsin genes. An electrophysiological study has confirmed that both L and M opsins contribute to the electroretinogram (ERG) response, i.e., gross electrical activity of the retina to light stimuli, in male and female howlers, indicating that both genes are expressed in both sexes (Jacobs et al. 1996). This ERG result also suggests that the L and M opsins of howlers have spectral sensitivities similar to those of the L and M opsins of catarrhine primates, i.e., λmax at about 562 and 530 nm (Jacobs et al. 1996). In a behavioral experiment, both male and female howlers were able to distinguish color hues that are difficult for dichromats to distinguish (Araujo et al. 2008). Based on these observations, howlers are considered to have evolved their “routine” trichromacy independently from catarrhine primates (Jacobs 2008).
What is not known, however, is whether trichromacy in howlers is as invariable as in nonhuman catarrhine primates. The loss of an L or M opsin gene and gain of L/M hybrid genes are major causes of color vision variation in humans, i.e., dichromacy and anomalous trichromacy, respectively (Deeb 2005), but are rarely found in other catarrhine primates (Hanazawa et al. 2001; Hiwatashi et al. 2011; Jacobs and Williams 2001; Onishi et al. 1999, 2002; Saito et al. 2003; Terao et al. 2005; Verrelli et al. 2008). If normal trichromacy leads to greater reproductive success, the incidence of dichromacy and anomalous trichromacy should be rare in howler populations. It follows that polymorphic color vision in most New World monkeys may be regarded as an intermediate stage of primate evolution from dichromacy to trichromacy, and if a gene rearrangement juxtaposing the L and M opsin genes occurs, it will quickly spread through population due to natural selection (Bowmaker et al. 1987; Jacobs et al. 1996). But if normal trichromacy is not always more adaptive than dichromacy and/or anomalous trichromacy, it would not be surprising to find these variants in howler populations arising by the relaxation of selective constraints maintaining normal trichromacy, or possibly by some selective advantage to the variants (Kawamura et al. 2012).
Thus, the examination of L/M opsin genetic variation in wild howler populations is of profound importance in our understanding of the evolution of both color vision polymorphism in platyrrhine primates, and routine and normal trichromacy in catarrhine primates. Here we contribute to this effort by examining the opsin genes of two species of wild howlers from diverse regions of Central America.
Methods
Sample Collection and DNA Extraction
The focal individuals are mantled howlers (Alouatta palliata) in Costa Rica (Santa Rosa Sector of the Área de Conservación Guanacaste) and Nicaragua (Maderas Rainforest Conservancy Biological Station, Ometepe), and Guatemalan (or Yucatan) black howlers (A. pigra) in Belize (Monkey River) (Table I). We collected fecal samples from 2009 to 2011 from one (CG), three (BE, PI, and VI) and five (AB, BR, CO, GR, and QU) social groups in Santa Rosa, Ometepe, and Monkey River, respectively (Table I). Because of the difficulty of individual identification, we did not genotype entire social groups. Rather, we analyzed one to three distinguishable individuals from each group.
We collected fecal samples using sterile cotton swabs and suspended the samples in 5 ml of ASL lysis buffer (QIAamp DNA Stool Mini Kit; Qiagen, Crawley, U.K.), pre-aliquoted in sterile 15-ml screw-capped plastic vials. Collectors wore a mask and plastic gloves to minimize the chance of contamination by humans. We stored the samples at ambient temperature until being shipped to Japan for extraction. We extracted DNA from fecal samples using the QIAamp DNA Stool Mini Kit following Hiramatsu et al. (2005).
The research and the sample export were permitted by Área de Conservación Guanacaste and the Ministerio de Ambiente y Energía (MINAE) of Costa Rica, the Maderas Rainforest Conservancy and the Molina family on Ometepe Island in Nicaragua, and local land owners in Monkey River, the Monkey River Tour Guide Association, and the Forest Department of Belize.
Determination of Entire Coding Sequence of L/M Opsin Genes
On the basis of the quantity of DNA recovered from fecal samples, we chose three individuals each of Alouatta palliata and A. pigra (IDs boldfaced in Table I) for nucleotide sequencing for all of the six protein-coding exons of the L/M opsin genes. We amplified the six exons from fecal DNA via polymerase chain reaction (PCR), using primers listed in Table SI. We designed these primers to obtain both L and M opsin sequences simultaneously by choosing noncoding regions conserved among L/M opsin genes of red howlers (Alouatta seniculus), spider monkeys (Ateles geoffroyi), and capuchins (Cebus capusinus) (Boissinot et al. 1997; Dulai et al. 1999; Hiwatashi et al. 2010).
We sequenced the intron 2 to determine the nucleotide haplotypes encompassing exon 2 through exon 3 in one of the three Alouatta palliata (CG-17) by “primer walking” with a series of overlapping PCR amplifications (Table SII and Fig. 1). We designed PCR primers to obtain both L and M opsin sequences simultaneously as we did with the exon PCRs. We took one primer sequence from the region where we determined the sequence for Alouatta palliata in this study (Table SII), and we designed the other primer from conserved sequence regions of 1) spider monkeys (Hiwatashi et al. 2010), 2) human genome project (GenBank accession nos. Z68193 and AC092402), and 3) the common marmoset (Callithrix jacchus) genome project [Ensembl Marmoset release 70, Genome assembly C_jacchus3.2.1 (GCA 000004665.1), X: 140700397~140702962].
PCR strategy for isolation of L and M opsin genes of Alouatta palliata and A. pigra and primer walking of intron 2 for A. palliata. The six exons of L/M opsin genes are depicted by solid boxes with exon numbers. An expanded view is given to show the primer walking strategy for intron 2. PCRed regions are indicated by double-headed arrows. In the expanded view, nucleotide sites that differ between L and M opsin genes are depicted by vertical bars.
We carried out PCRs in volumes of 50 μl containing 1.5 units of Ex Taq polymerase hot start version (Takara Biotechnology Co., Tokyo, Japan) with 1× Ex Taq buffer, 0.2 mM dNTPs, 2.0 mM MgCl2, 1 μM forward and reverse primers, and 5 μl of the DNA extract from feces. We set cycles at 94°C for 5 min followed by 40 cycles at 94°C for 30 s, at the annealing temperature indicated in Tables SI and SII for 30 s, and at 72°C for 30 s. Pure water was used as the template for the negative control in every reaction. We purified the amplified DNA fragments using UltraClean 15 DNA Purification Kit (MO BIO Laboratories, Inc., Carlsbad, CA).
We directly sequenced both strands of the purified DNA samples using Applied Biosystems model 3130 automatic sequencer with Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems Japan, Tokyo) and the PCR primers. We conducted the cycle sequencing reaction at 94°C for 5 min followed by 40 cycles at 94°C for 30 s, at 62°C for 30 s, and at 72°C for 2 min.
To determine the nucleotide haplotypes and distinguish sequences into L and M opsin types, we subjected the PCR-amplified DNA fragments to DNA cloning for all exons and intron 2 using pGEM-T vectors (Promega, Madison, WI). We sequenced them as described in the preceding text. We confirmed their nucleotide sequences by multiple PCR experiments. We distinguished L and M opsin sequences in exons 3, 4, and 5 according to the published sequences of Alouatta seniculus (Boissinot et al. 1997, 1998). We distinguished the two sequences of exon 2 in Alouatta palliata by haplotype extension from the exon 3 sequences based on primer walking. We distinguished the exon 2 sequences in Alouatta pigra according to their nucleotide similarity to the corresponding L and M sequences in A. palliata.
Phylogenetic Analysis
We estimated the number of nucleotide substitutions per site (d) for two sequences by the Jukes–Cantor method, and reconstructed the phylogenetic tree by applying the neighbor-joining method to the d values using the MEGA5 (Nei and Kumar 2000; Tamura et al. 2011). We evaluated the reliability of the tree topology by bootstrap analysis with 1000 replications.
“Three-Sites” Rule and Survey of the Exon 5 Sequence
The λmax value of primate L/M opsins can be estimated from the residue composition at the 180th, 277th, and 285th amino acid sites (“three-sites” rule) (Hiramatsu et al. 2004; Neitz et al. 1991; Yokoyama and Radlwimmer 1998, 2001; Yokoyama et al. 2008). The 180th site is encoded in exon 3 and the 277th and 285th sites are encoded in exon 5. Amino acid changes from L to M types at the three sites, from Ser to Ala at site 180 (denoted Ser180Ala), Tyr277Phe, and Thr285Ala, are expected to cause –5, –10, and –17 nm shifts of λmax, respectively (Yokoyama et al. 2008). Thus, the amino acids at the 277th and 285th sites explain the majority of the λmax difference between the L and M opsins. To describe the spectral variation of the L/M opsin genes in the howler populations, we surveyed exon 5 by PCR for all individuals in Table I, using the primer set for exon 5 (Table SI) under the same conditions described in the preceding text. We cloned and sequenced PCR-amplified DNA fragments as described to determine the 277/285 haplotypes. For males with recombinant 277/285 haplotypes, we further examined exon 3 using the primer set for exon 3 (Table SI).
Reconstitution of L/M Opsin Photopigments
We chemically synthesized DNA segments with the coding nucleotide sequences of the L and M opsin genes of Alouatta palliata and cloned them to vectors (Hokkaido System Science Co., Sapporo, Japan). We modified these clones so as to encode the same amino acid sequence of the M opsin of Alouatta pigra and a series of L/M hybrid opsins by site-directed mutagenesis using the QuickChange Site-Directed Mutagenesis Kit (Stratagene Japan, Tokyo). We sequenced all mutagenized DNAs to confirm that no spurious mutations were incorporated.
We cloned the opsin-coding DNAs into the pMT5 expression vector (Khorana et al. 1988) and transfected the clone into cultured COS-1 cells (RIKEN Cell Bank, Tsukuba, Japan). We incubated cells with 5 μM 11-cis retinal (Storm Eye Institute, Medical University of South Carolina, Charleston) and solubilized cells with 1% dodecyl maltoside. We purified the reconstituted photopigments using immobilized 1D4 antibody (University of British Columbia, Vancouver), as previously described (Hiramatsu et al. 2004; Kawamura and Yokoyama 1998). We recorded absorption spectra of the photopigments in 0.5-nm intervals, from 250 to 650 nm, using the U3010 dual beam spectrometer (Hitachi High-Technologies, Tokyo, Japan) at 20°C. We measured absorption spectra five times under dark conditions (dark absorption spectra). We measured five more times after exposing photopigments to a 60-watt room lamp light for 3 min (light absorption spectra) using Kodak Wratten Gelatin Filter No. 3, which cuts off wavelengths shorter than 440 nm. We subtracted the light absorption spectra from the dark absorption spectra to obtain the dark–light difference absorption spectra, with a negative peak at ca. 380 nm. The negative peak is caused by photoisomerization of 11-cis retinal to all-trans retinal, and its appearance is evidence of photosensitivity of the reconstructed opsins. We analyzed recorded spectra using UV Solution software (Hitachi High-Technologies).
Results
Determination of Entire Coding Sequence of L/M Opsin Genes
From the three individuals of Alouatta palliata and three of A. pigra for which we examined the entire coding sequences of the L/M opsin genes (Table I), we detected only one sequence for exons 1 and 6, and only two sequences for exons 2, 3, and 4 in each species after the cloning of PCR-amplified DNA fragments. We also detected two sequences for exon 5 in each species, except for one female of Alouatta palliata (CG-17) wherein we found an additional sequence. The entire coding sequences of the L and M opsin genes of the two species are shown in Fig. S1, together with the sequences of exons 3, 4, and 5 of Alouatta seniculus (the third exon 5 sequence from CG-17 is not included in Fig. S1). Between Alouatta palliata and A. pigra, the nucleotide sequences of the L opsin genes were identical, whereas the M opsin genes differed at two synonymous and one nonsynonymous sites (Fig. S1). We deposited these sequences in the DDBJ/EMBL/GenBank Data Libraries under accession nos. AB809459 (Alouatta palliata L opsin), AB809460 (A. palliata M opsin), AB809461 (A. pigra L opsin) and AB809462 (A. pigra M opsin).
In the phylogenetic tree reconstructed with other New World monkeys (Fig. 2), the L/M opsin gene sequences of Alouatta palliata and A. pigra clustered with the corresponding gene types of A. seniculus, demonstrating that the collected DNA sequences were indeed those of the study animals and were not the result of contamination. Within the Alouatta clade, A. seniculus was located outside of A. palliata and A. pigra in both L and M opsin genes. This is consistent with current knowledge of the phylogenetic relationships among Alouatta species (Cortes-Ortiz et al. 2003; Villalobos et al. 2004).
A phylogenetic tree reconstructed from entire coding nucleotides of L/M opsin genes from selected New World monkeys and humans. The L and M opsin genes of Alouatta palliata and A. pigra are highlighted with boldface letters. The bootstrap probabilities are given for each node. The scale bar indicates five nucleotide substitutions per 1000 sites. GenBank accession numbers: red howler (Alouatta seniculus) L opsin (AH007056) and M opsin (AH007055); white-faced capuchin (Cebus capucinus) L opsin (AB193772), I (intermediate) opsin (AB193778), and M opsin (AB193784); common marmoset (Callithrix jacchus) L opsin (AB046546), I opsin (AB046547) and M opsin (AB046548); black-handed spider monkey (Ateles geoffroyi) L opsin (AB193790) and M opsin (AB193796); human L opsin (Z68193) and M opsin (AC092402).
Survey of the Exon 5 Sequence
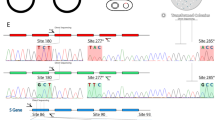
The third exon 5 sequence detected in one individual of Alouatta palliata (CG-17) was an M–L hybrid sequence with a recombinant 277/285 haplotype, Phe/Thr (Apa_ML in Fig. 3), between a normal L haplotype Tyr/Thr (Apa_L in Fig. 3) and a normal M haplotype Phe/Ala (Apa_M in Fig. 3). By extending the survey of the exon 5 sequence to all the other individuals in Table I, we examined a total of 26 X chromosomes from Alouatta palliata (N = 8) and A. pigra (N = 18). We found one male Alouatta pigra (CO-03) to possess an L–M hybrid sequence with a recombinant 277/285 haplotype, Tyr/Ala (Api_LM in Fig. 3). CO-03 also had Api_M but no normal L haplotype sequence (Table I). We also found the Api_LM exon 5 sequence in a female Alouatta pigra (AB-01) wherein we found Api_M together with an L haplotype sequence that contained M-type nucleotides at codons 274 and 275 (Api_L' in Fig. 3, Table I). We found this Api_L’ sequence in another male Alouatta pigra (BR-06) with Api_M (Table I). By sequencing exon 3 of the L/M opsin genes from CO-03 and BR-06, we found both L and M types of codons, encoding Ser and Ala, respectively, at site 180 in both individuals (data not shown). Assuming that the M type exon 3 sequences are from their Api_M, the L type exon 3 should be from Api_LM in CO-03 and Api_L' in BR-06. In the repeated PCR and cloning experiments, we observed the recombinant exon 5 sequences (Apa_ML, Api_LM and Api_L') from these individuals, but never detected recombinants in the others. There was no additional sequence detected in our survey and there were no insertion/deletion or nonsense nucleotide changes in the exons we sequenced. Thus, we did not find evidence that the howlers have any L/M opsin pseudogenes.
The exon 5 nucleotide sequences of Alouatta palliata L opsin (Apa_L), L–M hybrid opsin (Apa_ML), and M opsin (Apa_M); and A. pigra L opsin (Api_L), L opsin with M-type nucleotides at codons 274 and 275 (Api_L’), L–M hybrid opsin (Api_LM), and M opsin (Api_M). Identical nucleotides with Apa_L are indicated by dots. The codon positions 277 and 285 are indicated with L-type amino acids above Apa_L and M-type amino acids below Api_M.
Absorption Spectra
We reconstituted the L opsin photopigment common to Alouatta palliata and A. pigra and the M opsin photopigment for each of the two species. The λmax of the L opsin was 564 nm, and the M opsin was 532 nm for both species (Fig. 4). These values are close to previous estimates for the L and M opsins of Alouatta caraya and A. seniculus based on ERG measurements (562 and 530 nm for L and M opsins, respectively) (Jacobs et al. 1996). They are also concordant with predictions from the “three-sites” rule for the amino acid composition at sites 180, 277, and 285 (Ser/Tyr/Thr in L and Ala/Phe/Ala in M) for primate L/M opsins (560 and 532 nm, respectively) (Hiramatsu et al. 2004; Yokoyama et al. 2008).
Because the exon 3 sequence of Apa_ML was unknown, we synthesized two Apa_ML photopigments: one with Ser at site 180 (L-type exon 3) by introduction of Tyr277Phe into the L opsin expression construct common to Alouatta palliata and A. pigra, and the second with Ala at site 180 (M-type exon 3) by introducing Ala285Thr into the Apa_M. The λmax value of the former was 554 nm and the latter was 547 nm (Fig. 5a). The former is similar to an L/M opsin allele of a callitrichine [common marmoset at 553 nm (Kawamura et al. 2001)] and an ateline [spider monkey at 553 nm (Hiramatsu et al. 2008)], whereas the latter is similar to an L/M opsin allele of cebines [squirrel monkey at 545 nm (Hiramatsu et al. 2004) and capuchin at 543 nm (Hiramatsu et al. 2005)], measured by the same method. Because the exon 3 sequence of Api_LM was determined to be the L type, we introduced the Thr285Ala mutation into the common L opsin expression construct for reconstruction of the Api_LM photopigment. The λmax was similar to the L/M opsin allele of cebines, at 546 nm (Fig. 5b).
Absorption spectra of hybrid L/M opsins of Alouatta palliata and A. pigra. (a) Absorption spectra of two possible Apa_ML photopigments. Apa_L_Y277F was constructed by introducing Tyr277Phe into the L opsin common to the two species. Apa_M_A285T was constructed by introducing Ala285Thr into the M opsin of Alouatta palliata. (b) Absorption spectra of Api_LM photopigment constructed by introducing Thr285Ala into the common L opsin (Api_L_T285A). Insets show dark–light difference absorption spectra. The λmax values were taken directly from the dark absorption spectra.
Discussion
The λmax values of the reconstituted L and M opsin photopigments of the two howler species were 564 nm and 532 nm, respectively. This is equivalent to those found in catarrhine primates. We surveyed the exon 5 sequences encoding the diagnostic 277th and 285th residues for λmax for 8 and 18 X chromosomes from Alouatta palliata and A. pigra, respectively. We found one M–L hybrid sequence with a recombinant 277/285 haplotype Phe/Thr in Alouatta palliata and two L–M hybrid sequences with a recombinant haplotype Tyr/Ala in A. pigra. We also reconstituted the hybrid photopigments and showed that the λmax of the M–L hybrid opsin of Alouatta palliata was either 554 nm or 547 nm, and that of the L–M hybrid opsin of A. pigra was 546 nm. These are similar to the λmax values observed in polymorphic alleles of single-locus L/M opsins in other New World monkeys.
Assuming that the Alouatta palliata CG-17 (Table I) has the normal L (Apa_L) and the normal M (Apa_M) opsin genes on one X chromosome, as inferred from the other individuals of A. palliata, the M–L hybrid Apa_ML gene must reside on the other X chromosome with either Apa_L or Apa_M. However, the use of fecal DNA is problematic for estimating the linked gene type to Apa_ML, based on the difference of amplitude by PCR between Apa_L and Apa_M. Determination of the exon 3 sequence of Apa_ML by overlapping PCR is also improbable because we have not completed primer walking from exon 3 to exon 5, while distinguishing two genes, using fecal DNA. We will continue sampling populations of Alouatta palliata, in the hope of finding a male individual that possesses Apa_ML to determine the linked gene type to Apa_ML and the exon 3 sequence of Apa_ML, as in the case of Api_LM in A. pigra. Despite this, we are still able to determine a possible range of spectral separation between Apa_ML and the other L/M opsin in those males carrying them, based on the absorption spectra of their photopigments (Figs. 4 and 5a).
The spectral separation of λmax between Apa_ML and a normal L or M opsin could range from 10 nm (554 nm λmax of Apa_ML and 564 nm λmax of Apa_L), 15 nm (547 nm λmax of Apa_ML and 532 nm λmax of Apa_M), 17 nm (547 nm λmax of Apa_ML and 564 nm λmax of Apa_L), or 22 nm (554 nm λmax of Apa_ML and 532 nm λmax of Apa_M). In Alouatta pigra, we found Api_LM in a male with Api_M (CO-03 in Table I), and its λmax was 546 nm (Fig. 5b). Thus, in males carrying this X chromosome, the spectral separation between the two X-linked opsins is 14 nm. These ranges of spectral separation are comparable to those observed in trichromatic females carrying an intermediate-λmax allele of the single-locus triallelic L/M opsin gene in other New World monkey species (Hiramatsu et al. 2004, 2005; Kawamura et al. 2001) and trichromatic spider monkey females with a unique diallelic L/M opsin gene (Hiramatsu et al. 2008). These ranges also overlap with those observed in human anomalous trichromacy, wherein spectral separation is up to 12 nm in deuteranomaly (trichromatic color vision based on S, L, and an anomalous L-like photoreceptor) and up to 7 nm in protanomaly (based on S, M, and an anomalous M-like photoreceptor), but are larger overall (Deeb 2006). Thus, the anomalous trichromacy in howlers would be comparable to less severe anomalous phenotypes of human trichromats.
Amino acid changes Tyr277Phe and Thr285Ala are expected to cause larger spectral shifts to L/M opsins (–10 nm and –17 nm, respectively) than Ser180Ala (–5 nm) (Yokoyama et al. 2008). Thus, introduction of amino acid changes at 277 or 285 site would be subjected to more severe purifying natural selection than changes at 180. Indeed, in the case of human L/M opsin genes, the recombinant haplotype within exon 5 (between sites 277 and 285) is rare (Hayashi et al. 2006) and recombinant haplotypes are mainly between exon 3 (encoding the site 180) and exon 5 (Deeb 2005). In humans, the frequencies of protanomaly and deutanomaly by the aforementioned L/M hybrid opsins are ca. 1% and 4–5% in males, respectively, and the frequency of red-green color vision defects, including dichromacy and anomalous trichromacy, is ca. 4–8% in males of various geographic origins (Birch 2012; Deeb 2005). In nonhuman catarrhine primates, even the recombinant haplotype between exon 3 and exon 5 is reported to be absent or rare (Hanazawa et al. 2001; Hiwatashi et al. 2011; Jacobs and Williams 2001; Onishi et al. 1999, 2002; Saito et al. 2003; Terao et al. 2005; Verrelli et al. 2008). The frequency of L/M hybrid opsin genes in our samples of howlers was 12.5% (1 of 8 X chromosomes) in Alouatta palliata and 11.1% (2 of 18) in A. pigra. The high frequency of L/M hybrid opsins and their characteristic recombination between sites 277 and 285 contrasts greatly with catarrhine primates.
Further studies need to be conducted with larger sample sizes to determine the frequency of recombinant haplotypes, not only within exon 5 but also between exon 3 and exon 5. In addition, larger sample sizes should be surveyed to determine whether dichromacy (absence of one of the two L/M opsin genes on an X chromosome) occurs in howlers. Our study species are limited to the howlers inhabiting Central America. Further study of other howler species in South America is necessary to draw more general conclusions. However, our finding of high frequencies of different forms of L/M hybrid opsin genes in two howler species could imply a high prevalence of hybrid L/M opsins in other howler species. This finding challenges the current view that howlers, like nonhuman catarrhine primates, are routine and possibly uniform trichromats (Jacobs et al. 1996).
In humans, production of hybrid L/M opsin genes and deletion of L or M opsin gene are caused by unequal meiotic recombination and gene conversion (Deeb 2005; Shyue et al. 1994; Winderickx et al. 1993). Gene conversion replaces the nucleotide sequence of a part of one gene with the corresponding sequence of the other and lowers the local nucleotide difference between the two genes. A similar process could have created the hybrid opsin genes in howlers, although the juxtaposition of the L and M opsin genes in howlers occurred separately from the ancestor of the catarrhine primates as it not accompanied by duplication of LCR (Dulai et al. 1999). Introns are less susceptible to purifying selection than exons and are likely to represent better the occurrence of gene conversion. In our preliminary analysis of nucleotide difference between L and M opsin genes in introns, the nucleotide difference was higher in howler monkeys (Alouatta palliata) (1.0~1.8%; introns 2–4) than in human (0~0.3%; introns 1–5), chimpanzee (0.3%; intron 4) (Zhou and Li 1996), gibbon (1.0%; introns 3–4) (Hiwatashi et al. 2011), and baboon (0.9%; intron 4) (Zhou and Li 1996). This could infer a lower L–M gene-conversion rate in howlers than in catarrhine primates. Thus, the higher frequency of hybrid opsins in howlers could be attributed to other factors such as severity of selection. However, more detailed and comprehensive analysis of nucleotide sequence data is necessary to draw any conclusions on the difference of recombination/gene-conversion rate between howlers and catarrhine primates.
We found different trichromat phenotypes in howlers. Trichromacy in general is thought to be advantageous over dichromacy for detecting objects in red–green chromaticity that differ from the background foliage, especially from a long distance, most notably mature fruits, young leaves, pelage, and skin (Allen 1879; Bompas et al. 2013; Changizi et al. 2006; Dominy and Lucas 2001; Fernandez and Morris 2007; Lucas et al. 1998, 2003; Regan et al. 1998, 2001; Sumner and Mollon 2000, 2003; Surridge et al. 2003; Vorobyev 2004). Dichromacy, on the other hand, is advantageous in some visual tasks such as defeating cryptic coloration, motion detection, and foraging on surface-dwelling insects (Caine et al. 2010; Melin et al. 2007, 2010; Morgan et al. 1992; Saito et al. 2005b). In comparison, less is known about the relative abilities of different trichromat phenotypes. A rare protanomalous chimpanzee with a hybrid “R4G5” opsin (λmax at 538 nm) and a normal M opsin (λmax at 530 nm) was shown to be severely impaired in red–green chromatic discrimination, as were humans with protanomaly or deutanomaly (Saito et al. 2003), whereas anomalous trichromatic capuchins with a wider spectral separation of L/M opsin alleles (λmax at 545 nm and 530 nm) were successful in discriminating stimuli using Ishihara pseudo-isochromatic plates (Saito et al. 2005a). Conversely, in discriminating color-camouflaged stimuli, the chimpanzee performed as well as dichromats, whereas the capuchins were inferior to dichromats (Saito et al. 2005b). In a study of fig foraging behavior of free-ranging capuchins, trichromat monkeys with the most spectrally separated L/M opsin alleles (“normal” trichromats) showed the highest acceptance index for conspicuous figs, compared with anomalous trichromatic and dichromatic group mates, although there were no differences in feeding rates among phenotypes (Melin et al. 2009).
In nonhuman catarrhine primates, even mildly anomalous trichromats have not been found. This suggests that, at least in nonhuman catarrhine primates, mildly color-deficient phenotypes also suffer from some selective disadvantage. The higher frequency of anomalous trichromacy in howlers implies that the selective pressure to maintain “normal” trichromacy is lower in the Neotropics. However, in New World monkey species with polymorphic color vision, genetic studies have shown that the spectrally different alleles of the L/M opsin gene are actively maintained by balancing selection (Boissinot et al. 1998; Hiwatashi et al. 2010; Surridge and Mundy 2002). It is still an open question whether the selection is for 1) simply maintaining heterozygotes of L/M opsin alleles, i.e., trichromacy per se; 2) dichromacy and trichromacy; or 3) subtypic variation in dichromacy and/or trichromacy. It is known that normal trichromacy impedes some achromatic visual tasks such as breaking camouflage and motion detection, especially under low-light conditions (Kelber et al. 2003; Morgan et al. 1992; Perini et al. 2009). Further studies of howlers’ visual behaviors and the genetics of the L/M opsin variation will be beneficial in determining whether mildly anomalous trichromacy is more advantageous than normal trichromacy in achromatic tasks while maintaining the advantage in chromatic tasks, relative to dichromacy.
Whether or not the anomalous trichromacy in howlers has any selective advantage, its presence indicates that uniform and normal trichromacy is not necessarily the optimum result of color vision evolution in wild primates. It is still an open question as to whether the difference between nonhuman catarrhines (uniform trichromacy) and platyrrhines (polymorphic color vision) is attributable to a 1) biogeographic differences among continents, e.g., the severity of seasonality, or a prevalence of drably colored fruits and asynchronous species, e.g., figs and palm fruits (Dominy et al. 2003); 2) dietary variability, e.g., degree of dependence on insects, leaves, or colorful fruits and different food patch sizes (see Melin et al., 2013); 3) variation in social color signals (Changizi et al. 2006; Fernandez and Morris 2007). Further population-level studies of the L/M opsin genes and field observations of visual behaviors of howlers are important for elucidating the evolutionary forces acting on color vision polymorphism in platyrrhine primates and routine and normal trichromacy in catarrhine primates.
References
Allen, G. (1879). The color sense: Its origin and development. London: Trubner & Co.
Araujo, A. C., Didonet, J. J., Araujo, C. S., Saletti, P. G., Borges, T. R., & Pessoa, V. F. (2008). Color vision in the black howler monkey (Alouatta caraya). Visual Neuroscience, 25(3), 243–248.
Birch, J. (2012). Worldwide prevalence of red-green color deficiency. Journal of the Optical Society of America. A, Optics, image science, and vision, 29(3), 313–320.
Boissinot, S., Tan, Y., Shyue, S. K., Schneider, H., Sampaio, I., Neiswanger, K., et al. (1998). Origins and antiquity of X-linked triallelic color vision systems in New World monkeys. Proceedings of the National Academy of Sciences of the USA, 95(23), 13749–13754.
Boissinot, S., Zhou, Y. H., Qiu, L., Dulai, K. S., Neiswanger, K., Schneider, H., et al. (1997). Origin and molecular evolution of the X-linked duplicate color vision genes in howler monkeys. Zoological Studies, 36(4), 360–369.
Bompas, A., Kendall, G., & Sumner, P. (2013). Spotting fruit versus picking fruit as the selective advantage of human colour vision. i-Perception, 4(2), 84–94.
Bowmaker, J. K., Jacobs, G. H., & Mollon, J. D. (1987). Polymorphism of photopigments in the squirrel monkey: A sixth phenotype. Proceedings of the Royal Society of London B: Biological Sciences, 231(1264), 383–390.
Caine, N. G., Osorio, D., & Mundy, N. I. (2010). A foraging advantage for dichromatic marmosets (Callithrix geoffroyi) at low light intensity. Biology Letters, 6(1), 36–38.
Changizi, M. A., Zhang, Q., & Shimojo, S. (2006). Bare skin, blood and the evolution of primate colour vision. Biology Letters, 2(2), 217–221.
Cortes-Ortiz, L., Bermingham, E., Rico, C., Rodriguez-Luna, E., Sampaio, I., & Ruiz-Garcia, M. (2003). Molecular systematics and biogeography of the Neotropical monkey genus, Alouatta. Molecular Phylogenetics and Evolution, 26(1), 64–81.
Deeb, S. S. (2005). The molecular basis of variation in human color vision. Clinical Genetics, 67(5), 369–377.
Deeb, S. S. (2006). Genetics of variation in human color vision and the retinal cone mosaic. Current Opinion in Genetics & Development, 16, 301–307.
Dominy, N. J., & Lucas, P. W. (2001). Ecological importance of trichromatic vision to primates. Nature, 410(6826), 363–366.
Dominy, N. J., Svenning, J. C., & Li, W.-H. (2003). Historical contingency in the evolution of primate color vision. Journal of Human Evolution, 44(1), 25–45.
Dulai, K. S., von Dornum, M., Mollon, J. D., & Hunt, D. M. (1999). The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome Research, 9(7), 629–638.
Fernandez, A. A., & Morris, M. R. (2007). Sexual selection and trichromatic color vision in primates: Statistical support for the preexisting-bias hypothesis. The American Naturalist, 170(1), 10–20.
Hanazawa, A., Mikami, A., Sulistyo Angelika, P., Takenaka, O., Goto, S., Onishi, A., et al. (2001). Electroretinogram analysis of relative spectral sensitivity in genetically identified dichromatic macaques. Proceedings of the National Academy of Sciences of the USA, 98(14), 8124–8127.
Hayashi, T., Kubo, A., Takeuchi, T., Gekka, T., Goto-Omoto, S., & Kitahara, K. (2006). Novel form of a single X-linked visual pigment gene in a unique dichromatic color-vision defect. Visual Neuroscience, 23(3–4), 411–417.
Hiramatsu, C., Melin, A. D., Aureli, F., Schaffner, C. M., Vorobyev, M., Matsumoto, Y., et al. (2008). Importance of achromatic contrast in short-range fruit foraging of primates. PLoS ONE, 3(10), e3356.
Hiramatsu, C., Radlwimmer, F. B., Yokoyama, S., & Kawamura, S. (2004). Mutagenesis and reconstitution of middle-to-long-wave-sensitive visual pigments of New World monkeys for testing the tuning effect of residues at sites 229 and 233. Vision Research, 44(19), 2225–2231.
Hiramatsu, C., Tsutsui, T., Matsumoto, Y., Aureli, F., Fedigan, L. M., & Kawamura, S. (2005). Color-vision polymorphism in wild capuchins (Cebus capucinus) and spider monkeys (Ateles geoffroyi) in Costa Rica. American Journal of Primatology, 67(4), 447–461.
Hiwatashi, T., Mikami, A., Katsumura, T., Suryobroto, B., Perwitasari-Farajallah, D., Malaivijitnond, S., et al. (2011). Gene conversion and purifying selection shape nucleotide variation in gibbon L/M opsin genes. BMC Evolutionary Biology, 11(1), 312.
Hiwatashi, T., Okabe, Y., Tsutsui, T., Hiramatsu, C., Melin, A. D., Oota, H., et al. (2010). An explicit signature of balancing selection for color-vision variation in new world monkeys. Molecular Biology and Evolution, 27(2), 453–464.
Jacobs, G. H. (2008). Primate color vision: A comparative perspective. Visual Neuroscience, 25(5–6), 619–633.
Jacobs, G. H., Neitz, M., Deegan, J. F., & Neitz, J. (1996). Trichromatic colour vision in New World monkeys. Nature, 382(6587), 156–158.
Jacobs, G. H., & Williams, G. A. (2001). The prevalence of defective color vision in Old World monkeys and apes. Color Research and Application, 26(Supplement), S123–S127.
Kawamura, S., Hirai, M., Takenaka, O., Radlwimmer, F. B., & Yokoyama, S. (2001). Genomic and spectral analyses of long to middle wavelength-sensitive visual pigments of common marmoset (Callithrix jacchus). Gene, 269(1–2), 45–51.
Kawamura, S., Hiramatsu, C., Melin, A. D., Schaffner, C. M., Aureli, F., & Fedigan, L. M. (2012). Polymorphic color vsion in primates: Evolutionary considerations. In H. Hirai, H. Imai, & Y. Go (Eds.), Post-genome biology of primates (pp. 93–120). Tokyo: Springer.
Kawamura, S., & Yokoyama, S. (1998). Functional characterization of visual and nonvisual pigments of American chameleon (Anolis carolinensis). Vision Research, 38(1), 37–44.
Kelber, A., Vorobyev, M., & Osorio, D. (2003). Animal colour vision—behavioural tests and physiological concepts. Biological Reviews, 78(1), 81–118.
Khorana, H. G., Knox, B. E., Nasi, E., Swanson, R., & Thompson, D. A. (1988). Expression of a bovine rhodopsin gene in Xenopus oocytes: Demonstration of light-dependent ionic currents. Proceedings of the National Academy of Sciences of the USA, 85(21), 7917–7921.
Lucas, P. W., Darvell, B. W., Lee, P. K. D., Yuen, T. D. B., & Choong, M. F. (1998). Colour cues for leaf food selection by long-tailed macaques (Macaca fascicularis) with a new suggestion for the evolution of trichromatic colour vision. Folia Primatologica, 69, 139–154.
Lucas, P. W., Dominy, N. J., Riba-Hernández, P., Stoner, K. E., Yamashita, N., Loria-Calderon, E., et al. (2003). Evolution and function of routine trichromatic vision in primates. Evolution, 57(11), 2636–2643.
Melin, A. D., Fedigan, L. M., Hiramatsu, C., Hiwatashi, T., Parr, N., & Kawamura, S. (2009). Fig foraging by dichromatic and trichromatic Cebus capucinus in a tropical dry forest. International Journal of Primatology, 30(6), 753–775.
Melin, A. D., Fedigan, L. M., Hiramatsu, C., Sendall, C., & Kawamura, S. (2007). Effects of colour vision phenotype on insect capture by a free-ranging population of white-faced capuchins (Cebus capucinus). Animal Behaviour, 73(1), 205–214.
Melin, A. D., Fedigan, L. M., Young, H. C., & Kawamura, S. (2010). Can color vision variation explain sex differences in invertebrate foraging by capuchin monkeys? Current Zoology, 56(3), 300–312.
Melin, A. D., Hiramatsu, C., Parr, N. A., Matsushita, Y., Kawamura, S. & Fedigan, L. M. (2013). The behavioral ecology of color vision: Frugivory as a selective force. International Journal of Primatology, in press.
Morgan, M. J., Adam, A., & Mollon, J. D. (1992). Dichromats detect colour-camouflaged objects that are not detected by trichromats. Proceedings of the Royal Society of London B: Biological Sciences, 248(1323), 291–295.
Nei, M., & Kumar, S. (2000). Molecular evolution and phylogenetics. New York: Oxford University Press.
Neitz, M., Neitz, J., & Jacobs, G. H. (1991). Spectral tuning of pigments underlying red-green color vision. Science, 252(5008), 971–974.
Onishi, A., Koike, S., Ida, M., Imai, H., Shichida, Y., Takenaka, O., et al. (1999). Dichromatism in macaque monkeys. Nature, 402(6758), 139–140.
Onishi, A., Koike, S., Ida-Hosonuma, M., Imai, H., Shichida, Y., Takenaka, O., et al. (2002). Variations in long- and middle-wavelength-sensitive opsin gene loci in crab-eating monkeys. Vision Research, 42(3), 281–292.
Perini, E. S., Pessoa, V. F., & Pessoa, D. M. (2009). Detection of fruit by the Cerrado's marmoset (Callithrix penicillata): Modeling color signals for different background scenarios and ambient light intensities. Journal of Experimental Zoology, 311A(4), 289–302.
Regan, B. C., Julliot, C., Simmen, B., Vienot, F., Charles-Dominique, P., & Mollon, J. D. (1998). Frugivory and colour vision in Alouatta seniculus, a trichromatic platyrrhine monkey. Vision Research, 38, 3321–3327.
Regan, B. C., Julliot, C., Simmen, B., Vienot, F., Charles-Dominique, P., & Mollon, J. D. (2001). Fruits, foliage and the evolution of primate colour vision. Philosophical Transactions of the Royal Society of London B: Biological Sciences, 356(1407), 229–283.
Saito, A., Kawamura, S., Mikami, A., Ueno, Y., Hiramatsu, C., Koida, K., et al. (2005a). Demonstration of a genotype-phenotype correlation in the polymorphic color vision of a non-callitrichine New World monkey, capuchin (Cebus apella). American Journal of Primatology, 67(4), 471–485.
Saito, A., Mikami, A., Hasegawa, T., Koida, K., Terao, K., Koike, S., et al. (2003). Behavioral evidence of color vision deficiency in a protanomalia chimpanzee (Pan troglodytes). Primates, 44(2), 171–176.
Saito, A., Mikami, A., Kawamura, S., Ueno, Y., Hiramatsu, C., Widayati, K. A., et al. (2005b). Advantage of dichromats over trichromats in discrimination of color-camouflaged stimuli in nonhuman primates. American Journal of Primatology, 67(4), 425–436.
Shyue, S. K., Li, L., Chang, B. H., & Li, W.-H. (1994). Intronic gene conversion in the evolution of human X-linked color vision genes. Molecular Biology and Evolution, 11(3), 548–551.
Sumner, P., & Mollon, J. D. (2000). Catarrhine photopigments are optimized for detecting targets against a foliage background. The Journal of Experimental Biology, 203, 1963–1986.
Sumner, P., & Mollon, J. D. (2003). Colors of primate pelage and skin: Objective assessment of conspicuousness. American Journal of Primatology, 59(2), 67–91.
Surridge, A. K., & Mundy, N. I. (2002). Trans-specific evolution of opsin alleles and the maintenance of trichromatic colour vision in Callitrichine primates. Molecular Ecology, 11(10), 2157–2169.
Surridge, A. K., Osorio, D., & Mundy, N. I. (2003). Evolution and selection of trichromatic vision in primates. Trends in Ecology and Evolution, 18(4), 198–205.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739.
Terao, K., Mikami, A., Saito, A., Itoh, S., Ogawa, H., Takenaka, O., et al. (2005). Identification of a protanomalous chimpanzee by molecular genetic and electroretinogram analyses. Vision Research, 45(10), 1225–1235.
Verrelli, B. C., Lewis, C. M., Jr., Stone, A. C., & Perry, G. H. (2008). Different selective pressures shape the molecular evolution of color vision in chimpanzee and human populations. Molecular Biology and Evolution, 25(12), 2735–2743.
Villalobos, F., Valerio, A. A., & Retana, A. P. (2004). A phylogeny of howler monkeys (Cebidae: Alouatta) based on mitochondrial, chromosomal and morphological data. Revista de Biologia Tropical, 52(3), 665–677.
Vorobyev, M. (2004). Ecology and evolution of primate colour vision. Clinical and Experimental Optometry, 87(4–5), 230–238.
Winderickx, J., Battisti, L., Hibiya, Y., Motulsky, A. G., & Deeb, S. S. (1993). Haplotype diversity in the human red and green opsin genes: Evidence for frequent sequence exchange in exon 3. Human Molecular Genetics, 2(9), 1413–1421.
Yokoyama, S., & Radlwimmer, F. B. (1998). The "five-sites" rule and the evolution of red and green color vision in mammals. Molecular Biology and Evolution, 15(5), 560–567.
Yokoyama, S., & Radlwimmer, F. B. (2001). The molecular genetics and evolution of red and green color vision in vertebrates. Genetics, 158(4), 1697–1710.
Yokoyama, S., Yang, H., & Starmer, W. T. (2008). Molecular basis of spectral tuning in the red- and green-sensitive (M/LWS) pigments in vertebrates. Genetics, 179(4), 2037–2043.
Zhou, Y. H., & Li, W.-H. (1996). Gene conversion and natural selection in the evolution of X-linked color vision genes in higher primates. Molecular Biology and Evolution, 13(6), 780–783.
Acknowledgments
For local support, we thank R. Blanco Segura and the staff of the Área de Conservación Guanacaste, the Maderas Rainforest Conservancy, and the Molina family in Ometepe, and local forest guides and research assistants in Monkey River. We thank the following individuals for fecal sample collection and supplies: students in the Ometepe Primate Behavioral Ecology field courses of intersession 2010 and 2011 (Shana Wierchowski, Tom Kohlmann, Nate Thayer, Tom Nixon, Tom Kalnik, Monica Yoo, Lucas Fredericks, Ben Sapadin, Meagan Wheatley, Erin Steinwachs, Sandra Bender, and Julie Philipson), and students in the University of Calgary Research Methods field course (Alison Behie, Barb Kowalzik, Tracy Wyman, Kayla Hartwell, Jane Champion, and Brittany Dean) and Greg Bridget. We thank Fernando Campos for proofreading the manuscript and Amanda Melin and two anonymous reviewers for their comments. This study was supported by Grants-in-Aid for Scientific Research A 19207018 and 22247036 from the Japan Society for the Promotion of Science (JSPS) and Grants-in-Aid for Scientific Research on Priority Areas Comparative Genomics 20017008 and Cellular Sensor 21026007 from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S. Kawamura; SUNY Geneseo Research Foundation and the Anthropology Department to B. J. Welker; and the National Science and Engineering Research Council of Canada (NSERC), the National Geographic Society, the American Society of Primatologists, Conservation International, The International Primatological Society, Sigma XI, and the University of Calgary to M. S. Pavelka.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
The nucleotide sequences of Alouatta palliata L opsin (Apa_L), A. pigra L opsin (Api_L), A. seniculus L opsin (Ase_L), A. palliata M opsin (Apa_M), A. pigra M opsin (Api_M), and A. seniculus M opsin (Ase_M) genes. Identical nucleotides with Apa_L are indicated by dots. Sequences for exons 1, 2, and 6 of Ase_L and Ase_M have not been reported and are indicated by dashes. The deduced amino acid sequence of Apa_L is given above its nucleotide sequence by one-letter code. At sites where amino acids vary from Apa_L, they are indicated below the nucleotide sequence of Ase_M. The L-type and M-type amino acids at sites 180, 277, and 285 are seen in red and green, respectively. Boundaries between exons are indicated by vertical lines. Amino acid site numbers are indicated to the right of each line of the Apa_L amino acid sequence. (PDF 15.2 kb)
ESM 2
(PDF 882 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Matsushita, Y., Oota, H., Welker, B.J. et al. Color Vision Variation as Evidenced by Hybrid L/M Opsin Genes in Wild Populations of Trichromatic Alouatta New World Monkeys. Int J Primatol 35, 71–87 (2014). https://doi.org/10.1007/s10764-013-9705-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10764-013-9705-9