Abstract
The EU Water Framework Directive (WFD) aims to achieve good status of aquatic habitats. Classification of ecological status and identifying the stressors impacting aquatic habitats is essential for achieving this aim. Here, we evaluate different methods and indices for assessing ecological status in a hydropower-regulated river in central Norway using kick-sampling and macroinvertebrate identification through morphology and DNA as well as using environmental DNA. In Norway, the ASPT index (Average Score Per Taxon) is commonly used to evaluate ecological condition in general, although the index only provides evidence for organic pollution. We observed lower than expected diversity in the regulated river, but this was not reflected in the ASPT index, which showed “Good” to “High” status for all samples and methods. An alternative index, the IBIBI (Intercalibrated Benthic Invertebrate Biodiversity Index), returned “Bad” to “Moderate” status using the same data. The DNA-based identification methods returned in general higher species richness and somewhat higher index values than morphological species identification did. Our study exemplifies the importance of including relevant biological quality indices in WFD compliant assessments, and we advocate inclusion of a pressure-independent index like IBIBI in Norwegian river management and DNA-based identification methods for future river management in general.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent decades, biodiversity in freshwater systems (lakes, reservoirs, and rivers) has rapidly declined, both in terms of number of species and abundance of individual species (Reid et al., 2019). Several anthropogenic factors negatively impact biodiversity, and habitat degradation is a leading and persistent cause of population declines in freshwater ecosystems (Dudgeon et al., 2006). To prevent comprehensive deterioration, national water management legislations have urged for assessment and mitigation of the health of the river ecosystems in recent decades (e.g., Tharme, 1996; Dunbar et al., 2001; the U.S. Clean Water Act [Public Law 92-500]). In Europe, an ambitious aim is to achieve good ecological state of aquatic ecosystems through implementation of the EU Water Framework Directive (WFD) (European Commission, 2000).
The WFD defines multiple Biological Quality Elements (BQEs), targeting different taxonomic groups, to classify the ecological status of aquatic ecosystems. The status is based on deviation from a reference condition (expected natural status). This deviation is called the Ecological Quality Ratio (EQR) and that is translated into distinct classes of high, good, moderate, poor, or bad quality. The EQR is highly dependent on knowledge of the reference condition, for example, expected diversity of species present in an intact ecosystem (Reynoldson et al., 1997; Stoddard et al., 2006; Bongard et al., 2011). A well-established and defined reference condition allows for measurements of deviations from it (Reynoldson et al., 1997), and there should be a solid scientific basis for this definition, based either on knowledge from undisturbed sites, historical data, modelling data, or expert judgements (European Commission, 2000).
Benthic macroinvertebrates, which hold key ecological functions in the riverine ecosystem, and which are one of the BQEs of the WFD, are used for assessing the ecological status by estimating their diversity, taxonomic composition, abundance, and the ratio of sensitive to insensitive species (European Commission, 2000). In compliance with the WFD, several different macroinvertebrate indices have been used and developed to measure changes in environmental quality caused by different pressures, for example eutrophication, hydro-morphological deterioration, and acidification across Europe (Birk et al., 2012). One of the most commonly used benthic macroinvertebrate metrics is the Average Score Per Taxon index (ASPT) or its variant (Armitage et al., 1983; Birk et al., 2012; Jones et al. 2023). The ASPT index is made for providing evidence of organic pollution, with higher scores given to families known to be sensitive to oxygen depletion (Armitage et al., 1983), but it is often considered as a eutrophication or a general quality index (Norwegian Environment Agency, 2018; Swedish Agency for Marine and Water Management, 2018; Birk et al., 2018). The widespread use is understandable as excessive nutrient loading is a common environmental pressure affecting many rivers in Europe (European Environment Agency, 2019). Furthermore, the index is based on family-level identification, allowing most managers and consultants with limited taxonomic training to estimate this index and provide relevant data for assessing ecological status in relation to the WFD.
The WFD stipulates that member states should monitor those BQEs which are indicative of and most sensitive to the pressures to which each waterbody is subject (European Commission, 2000). In Norway, the standard guide to classification of environmental conditions in water (Norwegian Environment Agency, 2018) presents three different acidification indices and the ASPT index for benthic macroinvertebrates. Common practise is to calculate one of the acidification indices and the ASPT index when assessing ecological status in rivers (e.g., Thrane et al., 2020). However, these indices are not indicative of hydro-morphological pressures caused by hydropower operation. This shortcoming in the implementation of the WFD in Norway inspired us to study alternative indices.
One such indicator developed more than 10 years ago for Norwegian rivers, is the Intercalibrated Benthic Invertebrate Biodiversity Index (IBIBI) (Bongard et al., 2011). The IBIBI was developed to compensate for the poor performance of the ASPT index in Norwegian rivers and it considers all requirements for a robust and practical biomonitoring index as defined by Bonada et al. (2006). IBIBI is based on reference conditions (in a sense of European Commission, 2000) and an observed/expected species ratio for the three most identified orders of macroinvertebrates (EPT: Ephemeroptera, Plecoptera, Trichoptera), meaning that it can be used for assessing ecological status in relation to deviance from a reference condition (European Commission, 2000)—irrespective of pressure, given that an expected list of species for the specific region or ecotype exists (Bongard et al., 2018). It is based on species-level identification, which is the most reliable level for detecting environmental impacts, as species within the same family can have very different tolerance limits for different stressors (Resh & Unzicker, 1975; Resh & Rosenberg, 1993; Bongard et al., 2018). Like the ASPT index, IBIBI is based on presence only with no need for estimates of abundance (Bongard et al., 2011; Bongard et al., 2018). Lastly, it can be expanded to include other benthic invertebrate groups as well. However, IBIBI is not used in ecological status assessments today.
The potential for including any group of benthic invertebrates also makes the IBIBI very attractive for the inclusion of DNA-based identification methods in assessments in compliance with the WFD. Measurements of macroinvertebrate diversity are traditionally based on morphological analyses of samples collected using kick-sampling or Surber nets (Surber, 1937; Frost et al., 1971). These methods are well-tested and adopted in standardized methodology (e.g., Norwegian Environment Agency, 2018). However, sampling can be challenging in large river systems due to deep water and strong currents, and the morphological identification of each specimen is time consuming. A global decrease in the number of taxonomic experts further adds to the problem for large-scale monitoring projects (European Commission, 2022). Analyses based on DNA captured from organisms or from the environment (environmental DNA—eDNA) represent new and cost-effective methods for assessing macroinvertebrate biodiversity (Ruppert et al., 2019). These methods are scalable and can be used for processing many samples simultaneously, and they are less dependent on taxonomical expertise for the analyses of the data compared to morphological analyses. Importantly, DNA-based methods include species information on most taxa of macroinvertebrates in one single analysis and are not limited to groups of taxa which are possible to differentiate morphologically within a reasonable time frame. This enables the identification of many taxa in many samples in a relatively short time (Hering et al., 2018; Buchner et al. 2021), and the potential for representing broad-scale biodiversity in biological quality indices. Therefore, we wanted to test the applicability of DNA-based identification methods for macroinvertebrates in an ecological status assessment context, i.e., estimation of biodiversity and calculation of biological quality indices.
It is well-known that benthic macroinvertebrates are negatively impacted by reduction or modification of water flow caused by hydropower development (regulation hereafter) (e.g., Calapez et al., 2021). Regulation is in fact the most common pressure affecting rivers in Europe (European Environment Agency, 2019), and in Norway, hydropower regulation impacts approximately one third of all watercourses (Vann-Nett, 2022). Thus, status assessment in compliance with the WFD should also include evaluation of regulation. Here, we investigated how ecological status assessment of a regulated river was affected by choice of sampling material, identification method and biological quality index. We therefore compared results from sampling the organisms and identifying the taxa morphologically (morphology samples hereafter), with sampling the organisms and identifying the taxa using DNA extracted from the preservative ethanol (EtOH samples hereafter, e.g., Zizka et al., 2018) and by filtering water and identifying the taxa using eDNA extracted from water (water samples hereafter, e.g., Leese et al., 2021). We included all metazoas in our analyses but highlighted insects from the orders Ephemeroptera, Plecoptera and Trichoptera, which have traditionally been used for river assessments because they are sensitive to alterations of habitat or abiotic factors and they are abundant and easy to identify morphologically (Plafkin et al., 1989; Wallace et al., 1996; Bongard et al., 2018). Rather than finding a universal index for assessments of European benthic invertebrates, our primary aim was to demonstrate the possible bias in the generic use of the ASPT index in Norway and present an existing alternative (IBIBI) that could be adopted. Our secondary aim was to demonstrate that DNA-based identification methods are applicable in biodiversity and status assessments of benthic macroinvertebrates. Finally, we propose a series of recommendations for improved ecological status assessments of river benthic communities in general.
Material and methods
Study area
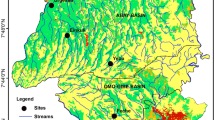
The river Nea is in central Norway (63° N 11° E, Fig. 1), and it is a heavily modified water body along its entire length (in a sense of European Communities, 2003). Thus, the river cannot achieve good ecological status, but good ecological potential given the hydro-morphological modification. This designation does not alter our results, but it is important for management of the river. However, we are referring to ecological status and not ecological potential throughout the paper for simplicity. The river originates from the Swedish lake Sylsjöen and drains approximately 80 km of mountain and forest areas into the lake Selbusjøen. In this study, we investigated a reach that covered approximately 29-km of the lowest part of the river, between the lakes Heggsetsjøen and Selbusjøen. There are three hydroelectric power plants operating in the studied part of the river; two started in 1962, and the third in 1989, and the study reach has been regulated for hydropower production since the 1980’s. The power stations exploit water from a river reservoir (Heggsetsjøen) about 36 km upstream of the river outlet. The water is led back into the river through a tunnel 10 km upstream of the river outlet. The minimum discharge between the dam and the tunnel outlet is 1.5 m3/s from May to September, while there are no specific discharge requirements in the other months. The mean water flow at the outlet in Lake Selbusjøen is approximately 70 m3/s. To compensate for reduced discharge and maintain water covered area, 34 weirs were constructed perpendicular to the flow direction in the mid-1980s, reducing the number of rapids in the river and creating new pools. The length of the weir pools and rapids are on average 426 m and 199 m, respectively, and the width of the river varies between 30 and 170 m based on measures in aerial photos (see Fig. 1 for an example). In the weir pools, water flow is very low and consequently the substrate is often covered by fine sediments (Sundt-Hansen et al., 2021). The river has been extensively studied in recent years in relation to river regulation and environmental design (i.e., Sundt-Hansen et al., 2021).
Location of sampling stations, weirs, and hydropower plants along a 32 km stretch of the river Nea in central Norway (a). Aerial photo of two sampling locations (red arrows), furthest upstream (b). Sampling stations were always chosen like this, in a rapid downstream of the weir and in a weir pool upstream of the weir where water was stationary. Weirs (blue arrow in b) and consecutive rapids are clearly visible in the photo, and the lengths of rapids and weir pools were estimated based on these photos. Photo from https://norgeibilder.no/
Field sampling and morphological identification
We collected benthic macroinvertebrate samples from 19 stations in June and October 2019 (Fig. 1), using a 500-µm kick-net. This aperture was used instead of 250-µm aperture to avoid excess debris in the samples. Smaller instars may have passed the net, but we expected to catch them when they had grown larger in the other sampling occasion based on over 30 years of sampling in the area (Aagaard et al., 2004). We chose sampling stations close to the weirs, where one station was downstream of the weir in a fast-flowing part of the river (rapids) and one station was upstream of the weir in a stationary part of each weir pool (see Fig. 1 for an example). We used kick-sampling instead of Surber sampling because the coarse substrate of Nea is unsuitable for Surber sampling. The samples were taken by holding the net downstream and kicking up the substrate while backing upstream. In stationary parts, the net was moved from side to side in front of the kicking feet to catch the material that was swirling up (Frost et al., 1971). We sampled different instream habitats proportionally to their existence at the stations following standardized methodology (Norwegian Environment Agency, 2018). We collected four-minute kick samples to reach reliable measures of biodiversity; estimating that we observed 95% of the taxa/station (Bongard et al., 2011). We picked the samples partly fresh and partly preserved in ethanol (Bongard et al., 2018) and all benthic groups were determined at the lowest possible taxonomic level and number of specimens was counted by an expert (Terje Bongard) with over 30 years of experience in benthic macroinvertebrate analyses in the area and using taxonomic literature (including Holmen, 1987; Lillehammer, 1988; Nilsson & Holmen, 1995; Engblom, 1996; Glöer, 2002; Rinne & Wiberg-Larsen, 2017). The taxonomic level of each taxon is given in the Online Resource 1 Table S2.
In addition to kick samples, we collected eDNA water samples from the same stations in June 2019. We collected water approximately 4 m from the riverside holding a 3-m pole and filtered 5 L through a 2.0 µm glass fibre filter (Merck, KGaA, Darmstadt, Germany), using a battery-powered peristaltic pump (Vampire sampler, Bürkle GmbH, Bad Bellingen, Germany). Immediately after filtration, we preserved the filters in 5-mL microcentrifuge tubes containing 4050 µl ATL-buffer (Qiagen GmbH, Hilden, Germany) and stored them at room temperature until DNA extraction.
DNA extraction, DNA metabarcoding and bioinformatics
To compare morphological identifications with DNA-based identification methods, we extracted total DNA from the ethanol preservative of the collected kick-net samples and from the water filter samples. Although analyses based on filtered ethanol show discrepancies with morphological analyses and may not be as precise as tissue-based analyses, ethanol-based analyses have proven to be a good method for aquatic insects and to record anthropogenic stressors (Zizka et al., 2018; Kjærstad et al., 2022). We filtered the ethanol through a 0.3-mm filter cloth, and then through a 0.45-µm cellulose nitrate filter (Nalgene Analytical Test Filter Funnels, ThermoFisher Scientific, Waltham, MA, USA), using a vacuum pump (Microsart e.jet, Sartorius GmbH, Göttingen, Germany) connected to a manifold (Pall Corporation, New York, NY, USA) carrying three individually operated filter holder bases. A smaller aperture was used for EtOH samples than for water samples due to practical reasons: 2.0 µm filter allowed more water to be filtered in the field while 0.45 µm filter was easier to handle in the laboratory. We do not expect this to cause large bias in our results (Turner et al., 2014). We placed the filters in 2-mL microcentrifuge tubes containing 1440 µL of ATL-buffer (Qiagen GmbH, Hilden, Germany) and added 160 µL of proteinase-K (Qiagen) prior to extraction. For DNA extraction of the eDNA water filters, we first added 450–500 µL of proteinase-K (Qiagen) to the 5-mL tubes and incubated them at 56 °C overnight. We extracted DNA from EtOH and water samples using the Nucleospin Plant II Midi kit columns (Macherey–Nagel GmbH, Düren, Germany) in combination with lysis and washing buffers from the Qiagen Blood & Tissue kit. We eluted DNA in 200 µL of pre-heated AE buffer (Qiagen) and re-eluted in the same column to maximize DNA yield.
For DNA metabarcoding, we amplified a 418 bp long fragment at the 3′ end of the standard mitochondrial COI DNA barcode region. The fragment was amplified with the freshwater macroinvertebrate targeting primers BF3 (Elbrecht et al., 2019) and BR2 (Elbrecht & Leese, 2017). The amplifications followed the two-step Illumina (San Diego, CA, USA) 16S protocol (Illumina, 2013). The PCR reactions had a final volume of 25 μL containing 2.5 μL DNA template (each template was diluted to have approximately 20 ng/µL of DNA), 12.5 μL 2 × KAPA HiFi HotStart ReadyMix (Merck KGaA, Darmstadt, Germany), 5 μL forward primer (2.5 µM), and 5 μL reverse primer (2.5 µM). All PCRs included negative control reactions (no DNA template). The PCR conditions were, with a heated lid, 94 °C for 3 min, followed by a total of 35 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 10 min. In the second PCR step, we dual-indexed Illumina-tailed amplicons, using Nextera DU (Illumina) indices under PCR conditions with a heated lid, 95 °C for 3 min, followed by a total of 8 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min. The second-step PCRs had a final volume of 50 µL containing 5 µL of the first-step PCR product, 25 µL 2 × KAPA HiFi HotStart ReadyMix, 5 µL of forward index, 5 µL of reverse index and 10 µL molecular grade H2O. We visualised the PCR products on a Tape Station (Agilent 4200, Agilent, Santa Clara, CA, USA) to check the amplification success and cleaned them with magnetic beads (MAGBIND RXN PURE PLUS, Omega Bio-Tek Inc., Norcross, GA, USA) after each PCR step. In the end, we normalised the indexed amplicons based on values from the Tape Station and pooled them into two libraries. The libraries were sequenced using the 600-cycle V3 Illumina MiSeq sequencing kit at the NTNU Genomics Core Facility in Trondheim, Norway.
We removed primers from the resulting raw amplicon reads using cutadapt v2.1 (Martin, 2011). Then we processed the reads with the R package DADA2 1.14 (Callahan et al., 2016) in R 3.5.3 (R Core Team, 2019). We adjusted the DADA2 parameters (Online Resource 1, Table S1) based on the quality profiles of the sequencing runs. We identified taxonomic affiliations of the generated ASVs (Amplicon Sequence Variants), using RDP classifier v 2.12 (Wang et al., 2007) with a trained reference database (Porter & Hajibabaei, 2018; downloaded September 17, 2019). In addition, we searched for highly probable Norwegian EPT species from the non-classified ASV data, using BLAST + 2.9.0 (Zhang et al., 2000), and added them to the results because the trained reference database was developed for North American use and evidently lacked some of the important local species. Finally, all EPT species assignments were manually checked against BOLD v.4 (Ratnasingham & Hebert, 2007).
Calculation of biological quality indices
We calculated ASPT index (Average Score Per Taxon; Armitage et al. 1983) values for morphology, EtOH and water samples based on the taxonomic assignments of all benthic macroinvertebrate taxa. The ASPT index is used for assessing ecological condition of rivers regarding eutrophication/organic load in Norway (Norwegian Environment Agency, 2018). It is based on the occurrence of a selection of higher taxa, mainly families, which are commonly found in running-water, and on a ranking of the various taxa's tolerance to organic load/nutrient salts. The index is intercalibrated, and the reference value for it is set to 6.9 for all water types in Norway (Norwegian Environment Agency, 2018). The class limits are set at 6.8 = high/good, 6.0 = good/moderate, 5.2 = moderate/poor and 4.4 = poor/bad. For making the ASPT index values comparable with other studies, we transformed them to nEQR (normalized Ecological Quality Ratio), using the transformation formula given in Norwegian Environment Agency (2018): nEQR = [(EQR − lower EQR class limit/higher EQR class limit − lower EQR class limit) × 0.2] + lower class limit where EQR is calculated by relating the ASPT index values to the reference value (original ASPT index value of 0 = 0 EQR and the reference value 6.9 = 1 EQR).
To relate the impact of regulation and weirs to natural state of the river, we followed publication of Bongard et al. (2011) and compiled a list of EPT taxa expected to be present in the river Nea assuming a natural ecological state, not taking hydropower regulation into account. This list is based on general knowledge of the biodiversity of river ecosystems in Norway and distribution patterns for EPT taxa in the region during the last 50 years (Nøst, 1986; Aagaard & Hågvar, 1987; Aagaard & Dolmen, 1996; Aagaard et al., 2002). The taxa were separated into three categories: commonly, sporadically, and rarely occurring (Online Resource 1, Fig. S4) as described in Bongard et al. (2011): “The species distributions and abundances of Ephemeroptera, Plecoptera and Trichoptera (EPT) follow a general three-way pattern: Some species are always rare regardless of distribution pattern, some species may be locally common, but rare over large areas, and some are common over large areas and regions.” and “Species predictions may be based on species distributions from regions, or in different river types within regions (classified by variables like size, altitude etc.). The topography, environment or size of these regions may of course vary considerable.”. The expected list of EPT taxa for the river Nea (Online Resource 1, Fig. S4) included 39 Ephemeroptera taxa, of which 20 were considered common, 12 sporadic, and 7 rare. For Plecoptera, the total number of expected taxa was 28 (25 common, 2 sporadic, and 1 rare), and for Trichoptera 96 (55 common, 16 sporadic and 25 rare). A more detailed description of the listing is available in the publication by Bongard et al. (2011).
Further, Bongard et al. (2011, 2018) suggested an index, the Intercalibrated Benthic Invertebrate Biodiversity Index (IBIBI), based on the common expected EPT species to be present in pristine running waters of the same kind within a specific region. The IBIBI is a deviance (EQR) from an expected common species list from its own region. Thus, the IBIBI expected common species list of a region may differ from an expected common species list of a given river (in our case, river Nea). This index is used partly in the Norwegian Nature index (including only Ephemeroptera and Plecoptera species, Certain et al., 2011), and it resembles RIVPACS approach and its derivates (Wright 2000; Aroviita et al., 2008). The main difference is that the IBIBI expected list of species is based on expert opinion while in RIVPACS approach, expected list of species is predicted based on models. The expected list of EPT taxa for the Central Norway IBIBI includes 27 taxa (Online Resource 1 table S3; Bongard et al., 2018), and the class limits are set at high > 22 taxa (“the taxonomic composition and abundance correspond totally or nearly totally to undisturbed conditions” from European Commission, 2000), good 18–22 (“there are slight changes in the composition and abundance of invertebrate taxa from the type-specific communities”), moderate 13–17 (“the composition and abundance of invertebrate taxa differ moderately from the type-specific communities”), poor 8–12 (“Waters achieving a status below moderate shall be classified as poor or bad.”) and bad < 8 (“Waters … in which large portions of the relevant biological communities normally associated with the surface water body type under undisturbed conditions are absent, shall be classified as bad.”). Here, we calculated the IBIBI based on the expected common EPT taxa observed in morphology, EtOH and water samples. Similar to the ASPT index, we calculated nEQRs (original IBIBI value of 0 = 0 EQR and the reference value 27 = 1 EQR), using the transformation formula (Norwegian Environment Agency, 2018).
Statistical analyses
We performed all statistical analyses in R, using the base R and the vegan v2.5–6 package (Oksanen et al., 2019). We compared the number of observed expected EPT taxa/sample, the number of observed EPT and Metazoa taxa in rapids and weir pools as well as ASPT and IBIBI index values based on morphology, EtOH and water samples, using non-parametric repeated measures tests (Friedman’s rank sum test across three sample types and Wilcoxon rank sum test for two sample types). In the case of post hoc analysis for the Friedman’s test, we adopted a code available at https://www.r-statistics.com/2010/02/post-hoc-analysis-for-friedmans-test-r-code/. We used non-parametric tests because variances were not homogenous (Levene’s test). In the case of species accumulation curves, we compared overlap of 95-% confidence intervals. In addition, we calculated Spearman’s correlation values for observed EPT species among the three sample types. We visualized the community compositions (EPT and Metazoa taxa) using nonmetric multidimensional scaling (NMDS) plots based on Bray–Curtis dissimilarity. To test differences in community compositions (EPT and Metazoa taxa), we calculated generalized discriminant analysis plots (CAP) based on a binomial (presence-absence) dissimilarity matrix and tested the significance of CAP analyses with permutation tests (999 permutations). We used this approach since it offers flexibility to choose an appropriate dissimilarity measure and it is powerful if the number of taxa observed is greater than the number of samples (Anderson & Robinson, 2003).
Results
In total, the kick samples included approximately 9800 EPT larvae, 4800 Chironomidae larvae, 1700 Simuliidae larvae and 800 specimens of other invertebrates (Odonata larvae, Coleoptera larvae, Mollusca, Clitellata, Acari and non-Chironomidae and non-Simuliidae Diptera larvae; Online Resource 1 Fig. S1). We collected on average 110 specimens/minute of kick-sampling, which is only approximately 18–22% of the expected number of specimens for the area (Bongard et al., 2011).
For the DNA metabarcoding, 3.8 million reads remained after quality control and merging the paired reads. Based on the reference sequences, we identified approximately 450 000 and 35 000 EPT reads in the EtOH samples and water samples, respectively (Fig. 2). In addition, we identified approximately 500 000 and 45 000 reads of class Insecta in EtOH and water samples, respectively, and 100 000 and 19 000 reads of the other target invertebrates in EtOH and water samples, respectively. The rest of the high-quality reads were either from non-target organisms (Fungi, Bacteria and unicellular Eukaryota; in total 1.1 million and 0.6 million reads in EtOH and water samples, respectively) or from Metazoa with affiliations at order level or higher (900 000 and 80 000 reads, of which 250 000 and 50 000 reads were affiliated with Insecta in EtOH and water samples, respectively).
Number of Metazoa and EPT (Ephemeroptera, Plecoptera and Trichoptera) taxa/sample in the morphology, EtOH and water samples collected in June (a). Number of taxa in different metazoan groups identified in the morphology, EtOH and water samples collected in June 2019 (b). Taxon accumulation curves for Ephemeroptera (c), Plecoptera (d), and Trichoptera (e) taxa based on morphology, EtOH, and water samples taken in June 2019. The shaded areas represent 95-% confidence intervals. The number of Metazoa species differ between kick-sample ethanol DNA and water eDNA samples (Wilcoxon signed rank test, P < 0.001). The number of EPT species differ among the morphologically identified kick samples and kick-sample ethanol DNA and water eDNA samples (Friedman rank sum test and following pairwise comparisons using Wilcoxon signed rank test, P < 0.001). Asterix (*) denotes the significant difference in (a)
The number of taxa and community composition using different approaches
We observed the most Metazoa taxa using water samples, followed by EtOH and morphology samples (Friedman rank sum test, P < 0.001, post hoc tests, P < 0.05; Fig. 2a). EtOH and water samples revealed rich communities of, for example, Diptera that we lumped at family-level in our morphological analysis (Fig. 2b, Online Resource 1 Table S2). Further, we found strong evidence that in total, we observed more EPT taxa using water samples than using morphology samples (Friedman rank sum test, P < 0.001, post hoc test for water and morphology, P < 0.001, and non-overlapping 95-% confidence intervals; Fig. 2c-e). However, we did not find evidence that the total number of EPT taxa differed using EtOH and morphology samples (post hoc test after Friedman’s test for water and morphology, P = 0.19, 95-% confidence intervals overlapped).
Although we detected similar numbers of EPT taxa in rapids and weir pools (Online Resource 1 Fig. S3), we found strong evidence that the EPT community composition was different based on both morphology and EtOH samples in rapids and weir pools (permutation test on generalized discriminant analysis results, P = 0.001; Online Resource 1 Fig. S4). However, there was no evidence of a difference in EPT community composition of the weir pools and rapids based on the water samples (permutation test on the results of the generalized discriminant analysis results, P = 0.34; Online Resource 1 Fig. S4).
Observed EPT diversity in relation to expected EPT diversity in the area
In total, we observed 91 (56%) of the 163 expected EPT taxa (Fig. 3a). The number of expected EPT taxa observed was higher in water and EtOH samples (73 and 68 taxa; 45% and 41%, respectively) than in morphology samples (54 taxa; 33%), and the DNA-based identification methods found more often the same taxa (all EPT in EtOH and water samples: Spearman’s rho = 0.44, P < 0.001, morphology and EtOH: rho = 0.30 P = 0.002, morphology and water: rho = 0.25 P = 0.01; Online Resource 1 Fig. S2). We observed a larger share of the common taxa (69%; Fig. 3b) compared to sporadic (38%; Fig. 3c) and rare taxa (33%; Fig. 3d). We observed 38 (55%) of the common expected species with all three identification methods, and the DNA-based identification methods found more often the same common taxa than morphology (EtOH and water samples: rho = 0.58, morphology and water: rho = 0.45, morphology and EtOH: rho = 0.39, P < 0.001 in all). In the case of sporadic and rare EPT species, the species found did not show any significant correlation among the methods (P > 0.05; Online Resource 1 Fig. S2).
A Venn diagram of observed and expected EPT taxa, using different sample types/identification methods in the whole dataset (a), common EPT taxa (b), sporadic EPT taxa (c) and rare EPT taxa (d). The expected and observed expected taxa are listed in Online Resource 1 Fig. S4. Proportions of expected taxa observed are depicted for Ephemeroptera (e), Plecoptera (f) and Trichoptera (g) in the whole dataset (June and October 2019). The proportions of observed to expected taxa are presented as proportions from total and as proportions from common, sporadic, and rare taxa. The box-plots present the observed taxa/sample and asterix (*) denotes a significantly different number of taxa/sample (Friedman’s rank sum test followed by pair-wise comparisons, P < 0.05)
The proportions of expected taxa observed varied substantially among the three orders (Fig. 3e-g): 40–70% of expected Ephemeroptera and Plecoptera taxa were observed while only 20–40% of expected Trichoptera taxa were observed, depending on the identification method.
On a per sample basis, we found strong evidence that the total number of expected EPT taxa observed per sample was the highest using water samples (Friedman’s rank sum test followed by pair-wise comparisons, P < 0.05, Fig. 3e-g). For common EPT taxa, number of observed taxa per sample followed the same pattern—the observed number was the highest using water samples (Friedman’s rank sum test followed by pair-wise comparisons, P < 0.05). In the case of sporadic and rare EPT species, morphology samples failed to find them except for two sporadic Ephemeroptera species (Online Resource 1 Fig. S2).
Biological quality indices
Despite the observed low abundance of EPT and the low number of observed expected EPT taxa, the ecological status of the study reach was considered “Good” when based on the ASPT index (average nEQR value 0.73, SD ± 0.11), and we did not find evidence that the ASPT index values differed among the identification methods (Friedman rank sum test P > 0.05, Fig. 4a-b).
Biological quality index values as normalised Ecological Quality Ratios (nEQR) in all stations using morphologically identified kick samples (Morphology) and kick-sample ethanol DNA (EtOH) and water eDNA (Water) samples. nEQRs are presented for aASPT in June, b ASPT in October, c IBIBI in June, and d IBIBI in October. The limit values of ecological status are given in horizontal lines. ASPT index values were similar using morphology, EtOH, and water samples in June (Friedman rank sum test, P > 0.05), and using morphology and EtOH in October (Wilcoxon rank sum test, P > 0.05). IBIBI index values using water samples differed from morphology and EtOH samples in June (Friedman rank sum test, P < 0.001, post hoc test P < 0.001 and P = 0.002, respectively), and using morphology and EtOH samples in October (Wilcoxon rank sum test, P < 0.001). The average index values for each method are given as + signs, and asterix (*) denotes significant differences among the methods
However, the ecological status of the study reach was considered “Poor” when based on the IBIBI (average nEQR value 0.28, SD ± 0.17, Fig. 4c-d). Moreover, we found strong evidence that the IBIBI values differed among the identification methods. They were the highest using water samples in June (Friedman rank sum test, P < 0.001, post hoc test for water and morphology, P < 0.001 and for water and EtOH, P = 0.002; Fig. 4c), and higher using EtOH than morphology samples in October (Wilcoxon rank sum test, P < 0.001; Fig. 4d). In fact, these differences lead to a quite different conclusion of the ecological status of the study reach: if based on the average IBIBI of water samples in June it is considered “Moderate” while if based on EtOH and morphology samples it is considered “Bad”.
Discussion
This study was based on sampling macroinvertebrates spring and autumn from 19 stations along a 29-km reach of a hydropower-regulated Norwegian river and identifying macroinvertebrates using both morphology and DNA as well as calculating two different biological quality indices for the study reach. DNA metabarcoding holds great potential for increasing the monitoring effort without increasing analytical costs linearly (Fediajevaite et al., 2021). This is of importance since recovery of freshwater biodiversity has halted and there is a need to characterise and evaluate status of aquatic communities more flexibly than previously (Haase et al., 2023). Here, our aim was to contribute to both fields—showing applicability of DNA metabarcoding and finding flexible status assessment tools. Considering our DNA metabarcoding results, they revealed that water eDNA samples had superior species detection rates, followed by DNA metabarcoding of EtOH from kick samples and lastly morphological analyses of kick samples. However, water samples failed to detect differences in community composition over the short distances between rapids and weir pools that were detected with morphology and EtOH samples, consistent with earlier studies (Macher et al., 2018; Hajibabaei et al., 2019; Gleason et al., 2021), and reflecting the fact that water samples may cover diversity upstream (Deiner et al., 2016). All methods detected the common expected EPT taxa relatively well, but morphology failed to find sporadically and rarely occurring expected species. Considering our status assessment results, the studied reach of the river was in good ecological status based on the ASPT index using all identification methods. In contrast, the status assessment based on the IBIBI varied from moderate to bad depending on analysis and time of sampling indicating that IBIBI was able to measure the regulation or weir construction-impaired status of the study reach.
Performance of sampling and identification methods
Representative samples are a prerequisite for reliable assessments of the diversity and ecological status of rivers. Kick-net sampling of benthic macroinvertebrates has some uncertainty related to the sampled substrate—the methodology is developed for loose stony substrate (like in river Nea), and in areas with large stones and blocks or sand and clay it is more difficult to catch the specimens to the net when kicking (Frost et al., 1971). Thus, sampling method needs to be adjusted according to the substrate (European Commission, 2000). However, even large kick samples cover only small areas of the riverbed in practice—individuals of a rare species can be several tens of meters apart, so the probability of registering such a species is very low (Aagaard et al., 2004; Bongard et al., 2011). In this sense, it is not surprising that our morphology samples failed to find sporadically and rarely occurring expected EPT species. That is also why biological quality indices based on expected and observed taxa lists consider usually only common species (Aroviita et al., 2008; Bongard et al, 2011).
Water eDNA sampling of benthic invertebrates resembles kick-net sampling to some degree—both methods are collecting the target from water and not from their actual habitat. However, eDNA sampling is based on collecting traces shed by the specimens from the environment while kick-sampling is based on disturbing the habitat of the specimens and collecting them. This means that the spatial scale of kick-net and eDNA sampling differs substantially; kick nets collect from local (micro-)habitats while eDNA samples may represent all habitats some distance upstream in the river (Deiner et al., 2016). Further, capturing eDNA from water is dependent on factors like pore size and type of filter, pH and organic and inorganic particles in the water, eDNA degradation rate, DNA shedding rate, behaviour of the species, mixing of water, etc. (reviewed in e.g., Goldberg et al., 2016; Bruce et al., 2021). In theory this means that with eDNA we could cover up to several kilometres of the upstream river habitats and get a comprehensive view of the diversity within that larger area (Pont et al., 2018), also including the rarest species due to both the spatial coverage of eDNA sampling and the sensitivity of the DNA metabarcoding (Balasingham et al., 2018; Leese et al., 2021), but in practice, this is rarely true (Hajibabaei et al., 2019).
Contrary to several other studies (e.g., Macher et al., 2018; Hajibabaei et al., 2019; Gleason et al., 2021) but consistent with the theoretical aspect of the eDNA sampling, our water samples detected more metazoan and EPT taxa important for regulatory biomonitoring than EtOH and morphology samples. However, since the distance between adjacent rapid and weir pool samples was only 100–200 m, water samples did not capture the community differences between the rapids and the weir pools, which were captured by the kick samples, as expected. Further, our results indicated that there is little difference whether specimens in kick samples were identified based on morphological characters or based on DNA metabarcoding of preservative ethanol. There are several reasons that may cause differences in taxa lists, either in morphology- or in DNA-based methods, and we try to summarise some obvious reasons and solutions to these issues below.
Small instars, eggs, pieces of dead specimens and eDNA may contribute DNA in the kick-net and water samples but are difficult or impossible to identify by conventional methods, which may be reflected in our results. Kick sampling both spring and autumn increases the probability of identifying the small instars for overall biodiversity assessment (Aagaard et al., 2004) but does not resolve the presence of dead organismal or environmental DNA in kick-net samples. This may lead to somewhat increased species detection rates and must be considered when using DNA metabarcoding as an identification method—this is why recalibration of existing biological quality indices may be needed (Vitecek et al., 2021).
PCR inhibition is a common issue when dealing with samples containing non-target organic matter (Albers et al., 2013). It may lead to poor amplification success and following sequencing results. Here, we did not specifically test samples for inhibition, but based on our amplification and sequencing results we do not expect inhibition having a significant effect in our results. If detected, inhibition can be reduced for example by diluting DNA extracts or by an extra column purification step in the DNA extraction phase (McKee et al., 2015).
Another PCR-related factor that certainly affects results is the suitability of the primers for target organisms (Maher et al., 2018; Leese et al., 2021). A related issue, but less severe, is the species-specific bias in the primer binding efficiency (Elbrecht & Leese, 2015). The choice of primers is of considerable concern since it may lead to false negatives and contribute to significantly lower diversity and ecological quality assessment results than expected. There is a trade-off between specificity and detection when choosing the primers to be used—the more general the primers are the broader diversity they will detect, but also the more non-target organisms and genomic regions will populate the results. In our case, the universal BF3 and BR2 primers (Elbrecht & Leese, 2017; Elbrecht et al., 2019) which have been found to amplify excessively the non-target organisms especially when applied to eDNA samples (Maher et al., 2018), worked equally or even better for water samples than for kick samples. We can speculate that our water samples did not contain as much non-target DNA or other confounding elements as water samples in studies that have suffered from poor target amplification (Maher et al., 2018; Leese et al., 2021). The poor performance of the general primers has led to a successful development of more specific primers targeting freshwater invertebrates in water samples (Leese et al., 2021). Another solution for the primer bias is to use multiple primers, which increases detection rate of species but also analytical costs (Zhang et al., 2018). Since the choice of primer is of paramount importance for acquiring representative results of diversity, there is a need for coherence, if not standardisation, of primers used in the WFD implementation and in environmental management in general.
Incomplete reference sequence libraries have been a persistent impediment when considering application of DNA metabarcoding in the environmental management context (Piper et al., 2019). If there is no reference sequence of a given species, identification based on DNA metabarcoding will either ignore or misidentify the species. However, the continuous population of the reference libraries has increased the coverage of the libraries (Weigand et al., 2019). In our case, all morphologically identified EPT species had a reference sequence available, and we did not suffer from incomplete libraries in this sense, but since we used a North American reference library (Porter & Hajibabaei, 2018) as a starting point, we initially missed some European species. However, we were able to correct the results based on our blast and BOLD searches. This exemplifies the importance of choosing and using reference libraries that match the sampled environment and the question to be answered (Mugnai et al., 2023).
In conclusion, we can advocate using DNA-based identification of specimens in kick samples since the difference to conventional morphological identification method did not influence much the assessment of biodiversity or ecological status of the river Nea. However, proper inclusion of the method in the WFD implementation may require redefinition and recalibration of reference conditions and indices using these techniques as suggested by Vitecek et al. (2021). DNA-based identification is especially beneficial if considering not only EPT but all benthic invertebrate taxa. The ability to identify all metazoans to species-level using DNA is very intriguing as they include many widespread and common species that could be highly relevant for water-quality monitoring (Sæther, 1979). Additional benefits arise if the number of samples is beyond capacity of personnel identifying specimens based on morphological characters and/or if DNA-based results can be controlled by doing conventional analyses for a subset of samples. In the case of eDNA, we can conclude that the methodology still needs development and the current benthic invertebrate indices for WFD compliant assessments may not be adequate for inclusion of eDNA. Thus, eDNA indices should be developed as suggested earlier (e. g., Vitecek et al., 2021), and we advocate following the principles of Bonada et al. (2006) in this work.
Performance of biological quality indices
Ecological quality assessment according to the WFD is a classification operation, which combines information from biological, hydro-morphological, physical and chemical elements (European Comission, 2000). Evaluation of status of benthic invertebrate communities through diversity and biological quality indices is only a part of this process, but because of their sensitivity to alterations of habitat and abiotic factors (Plafkin et al., 1989; Resh & Rosenberg, 1993; Wallace et al., 1996; Graf et al., 2008, 2009), benthic macroinvertebrates are important and good indicators of environmental quality. This is why considerable amount of research has been invested to develop, test, and calibrate different indices (reviewed in e. g., Birk et al., 2012; Vitecek et al., 2021).
In line with the WFD, Norway has implemented standardized methods and tools to assess ecological status of rivers (Norwegian Environment Agency, 2018), but the current ecological status assessment is only evaluating effects of acidification and organic pollution. This does not comply with the requirements of the WFD, which demands member states to monitor the pressures to which each waterbody is subject (European Commission, 2000). This may lead to an incorrect status assessment if the studied river is impacted by other environmental pressures such as hydro-morphological deterioration or by several pressures in concert (Jones et al., 2023). Norway also participated in the European intercalibration exercises with the ASPT index (Van de Bund, 2009; Birk et al., 2018). However, Norway has adopted a single reference value for both rivers and lakes in the whole country while for example Sweden, with similar water types, uses three different reference values for lakes (Swedish Agency for Marine and Water Management, 2018). This shortcoming has been discussed in the Norwegian river monitoring program and it is stated that at least humic rivers and naturally nutrient-poor high-altitude rivers should have less strict class boundaries (e. g., Thrane et al., 2020). In fact, the WFD demands type-specific reference values (European Commission, 2000) and a single reference value for all types of waterbodies negates the purpose of using a reference-based approach.
While this shortcoming in the Norwegian application of the ASPT index most probably did not skew our evaluation of organic pollution/eutrophication status of river Nea—the ASPT index showed “Good” status of the study reach in line with, for example, nutrient concentrations that have shown “High” status earlier (data from year 2007; Vann-Nett, 2022)—it may have implications for general ecological status assessments and their understanding. Since the ASPT index is intercalibrated and reported in nEQRs, it is easy to comprehend and use. At the same time, diversity should be reported and included in the status assessments. However, it is not reported as nEQRs in Norway and may easily be ignored when water managers are reading the assessments and planning for actions. In our case, diversity of EPT is lower than expected, and therefore, IBIBI is showing “Poor” to “Moderate” condition likely caused by the regulation (average water flow is 15–60% of natural condition; Online Resource 1 Fig. S5) and/or the many constructed weirs, but this information might be ignored if following the national guidance where acidification and eutrophication are the main stressors monitored (Norwegian Environment Agency, 2018).
That said, we would like to draw attention to the IBIBI as an existing metric that is relatively easy to incorporate in the national guidelines. The IBIBI was developed to compensate for the poor performance/implementation of the ASPT index in Norway (Bongard et al., 2011), considering the requirements for a robust and practical index suggested by Bonada et al. (2006). Importantly, the IBIBI is, in contrast to the ASPT index, based mostly on species-level identification. It is well-known that species within the same family can have different tolerance limits for different kinds of stressors (Resh & Rosenberg, 1993; Resh & Unzicker, 1975; Bongard et al., 2018). Losing a sensitive species of a given family due to regulation for example does not decrease the ASPT index value if a regulation-tolerant species still exists in an impacted site. Since IBIBI is based on observed to expect species ratios it can be used for assessing tolerance for all forms of perturbations and in different regions and water types given that an expected list of species exists (Bongard et al., 2018). Lastly, it can be expanded to include any other benthic invertebrate group as well, for example chironomids and oligochaetes that show wide applicability in impact assessments (Sæther, 1979).
Indices based on observed to expect species ratios, resembling IBIBI in that sense, are already in use in several European countries (Wright 2000; Davy-Bowker, et al., 2006; Aroviita et al., 2008). These RIVPACS-type predictive models use discriminant analysis to derive expected species lists of reference conditions and observed species lists of test sites are related to the expected species lists by calculating observed/expected (O/E) ratios for biotic indices. These are equivalent to the EQRs of the IBIBI, and importantly, of the WFD (European Commission, 2000). By including IBIBI in the assessment guidelines (Norwegian Environment Agency, 2018), Norway would comply to the WFD requirements, strengthen its ecological status assessment tools and would be able to contribute more broadly in intercalibration of the WFD compliant assessment tools in the future.
Conclusion
The EU WFD stipulates that member states should aim to reach good status of all surface waters by 2027 (European Commission, 2000). Regulation of water levels in rivers is one of the most common anthropogenic factors that degrade ecological status, which is a key proxy for the overall assessment of environmental quality in the WFD. We find it worrying that the index used for assessing ecological status in Norway,, the ASPT index, shows good ecological conditions in a heavily regulated ecosystem where only one-fifth of the expected abundance and half of the expected diversity of EPT taxa are present. However, implementing an organic pollution index for assessing hydro-morphological pressures does not comply with the requirements of the WFD. We show that an alternative biological quality index, the IBIBI, captures this impact, and therefore, should be considered for future impact assessments of regulation. In addition, we show that DNA-based identification increases taxon resolution, and that choice of sample (kick-sample or eDNA water sample) must be proportionate to the scale of the area monitored. The use of eDNA extracted from water may contribute to a larger-scale monitoring, while kick-sampling accompanied with morphology or DNA-based analysis is superior for detecting changes over shorter scales.
In the end, we would like to draw one specific recommendation from our investigations to the Norwegian river management authorities and four recommendations to the European water management in general. (1) Strictly speaking, the ASPT index only assesses organic pollution and other indices are needed to assess other stressors and ecological status in general. As an example, we showed that the ASPT index fails to detect effects of regulation, which is one of the most common pressures affecting Norwegian rivers. We recommend to further develop and incorporate the IBIBI in the national guidelines to detect any kind of pressure. IBIBI only covers two regions in its current form, and therefore, IBIBI status classes should be evaluated and expanded to cover the whole of Norway. Further, IBIBI should be expanded to cover taxa beyond EPT for a wider taxonomic assessment in water management. (2) In line with earlier studies (summarised in Bruce et al., 2021), our results show that DNA-based species identification has matured to be a suitable method for WFD compliant species richness assessment. Therefore, continued efforts should be made to incorporate DNA-based species identification as an alternative for ecological status assessment of benthic macroinvertebrate communities. DNA-based species identification methods can provide diversity assessments and estimate indices measuring various stressor effects, potentially with recalibration of the class boundary values due to increased species detections using DNA. (3) More focus should be put on the sampling methodologies in the WFD compliant assessments since the key for a reliable status assessment is representative samples, irrespective of the downstream species identification techniques. (4) Our results corroborate earlier findings that kick-net sampling is superior when detecting changes in local communities while water eDNA sampling is covering a larger area upstream. This should be considered if incorporating capture of water eDNA as a sampling method in WFD compliant assessments. There is still need for studies and standardisation of the eDNA sampling methodology. (5) Since the eDNA sampling is different from the standard benthic macroinvertebrate sampling, new metrics need to be developed to be applicable in WFD compliant assessments. This index development should follow the general principles to advance the indices towards “ideal” biomonitoring tools (Bonada et al., 2006).
Data availability
Data available from the ENA SRA repository with study name PRJEB49169. Occurrence data are available at GBIF DOI: https://doi.org/10.15468/c9rsg7.
References
Aagaard, K., T. Bækken & B. Jonsson, 2002. Felles instituttprogram. Virkninger av forurensninger på biologisk mangfold: Vann og vassdrag i by- og tettstedsnære områder. Sluttrapport 1997-2001. NINA Temahefte 19, Trondheim, Norway.
Aagaard, K. & S. Hågvar, 1987. Sjeldne insektarter i Norge 1. Økoforsk utredning nr. 6. Program for anvendt økologisk forskning. Økoforsk, Trondheim, Norway.
Aagaard, K. & D. Dolmen, 1996. Limnofauna Norvegica. Tapir forlag, Trondheim, Norway.
Aagaard, K., J. O. Solem, T. Bongard & O. Hanssen, 2004. Studies of aquatic insects in the Atna River 1987–2002. Hydrobiologia 521: 87–105. https://doi.org/10.1023/B:HYDR.0000026352.40631.37.
Albers, C. N., A. Jensen, J. Bælum & C. S. Jacobsen, 2013. Inhibition of DNA polymerases used in Q-PCR by structurally different soil-derived humic substances. Geomicrobiology Journal 30: 675–681. https://doi.org/10.1080/01490451.2012.758193.
Anderson, M. J. & J., Robinson, 2003. Generalized discriminant analysis based on distances. Australian & New Zealand Journal of Statistics 45: 301–318. https://doi.org/10.1111/1467-842X.00285.
Armitage, P. D., D. Moss, J. F. Wright & M. T. Furse, 1983. The performance of a new biological water-quality score system based on macroinvertebrates over a wide-range of unpolluted running-water sites. Water Research 173: 333–347. https://doi.org/10.1016/0043-13548390188-4.
Aroviita, J., E. Koskenniemi, J. Kotanen & H. Hämäläinen, 2008. A priori typology-based prediction of benthic macroinvertebrate fauna for ecological classification of rivers. Environmental Management 42: 894–906. https://doi.org/10.1007/s00267-008-9173-8.
Balasingham, K. D., R. P. Walter, N. E. Mandrak & D. D. Heath, 2018. Environmental DNA detection of rare and invasive fish species in two Great Lakes tributaries. Molecular Ecology 27: 112–127. https://doi.org/10.1111/mec.14395.
Birk, S., W. Bonne, A. Borja, S. Brucet, A. Courrat, S. Poikane, A. Solimini, W. van de Bund, N. Zampoukas & D. Hering, 2012. Three hundred ways to assess Europe’s surface waters: an almost complete overview of biological methods to implement the Water Framework Directive. Ecological Indicators 18: 31–41. https://doi.org/10.1016/j.ecolind.2011.10.009.
Birk, S., J. Böhmer & F. Schöll, 2018. Intercalibrating the national classifications of ecological status for very large rivers in Europe – Biological quality element – Benthic invertebrates. European Commission Joint Research Centre. https://data.europa.eu/doi/https://doi.org/10.2760/443119
Buchner, D., T.-H. Macher, A. J. Beermann, M.-T. Werner & F. Leese, 2021. Standardized high-throughput biomonitoring using DNA metabarcoding: Strategies for the adoption of automated liquid handlers. Environmental Science and Ecotechnology 8: 100122. https://doi.org/10.1016/j.ese.2021.100122.
Bonada, N., N. Prat, V. H. Resh & B. Statzner, 2006. Developments in aquatic insect biomonitoring: a comparative analysis of recent approaches. Annual Review of Entomology 51: 495–523. https://doi.org/10.1146/annurev.ento.51.110104.151124.
Bongard, T., O. H. Diserud, O. T. Sandlund & K. Aagaard, 2011. Detecting invertebrate species change in running waters: an approach based on the sufficient sample size principle. Bentham Open Environmental & Biological Monitoring Journal 4: 72–82. https://doi.org/10.2174/1875040001104010072.
Bongard, T., K. Johansen & T. B. Munkeby, 2018. A New Sampling Protocol and Intercalibrated Index for Invertebrates in Running Water. NINA Report 1548. Norwegian Institute for Nature Research, Trondheim, Norway.
Bruce, K., R. Blackman, S. J. Bourlat, A. M. Hellström, J. Bakker, et al 2021. A practical guide to DNA-based methods for biodiversity assessment. Pensoft Advanced Books, Sofia, Bulgaria. https://doi.org/10.3897/ab.e68634
Calapez, A. R., S. R. Q. Serra, R. Rivaes, F. C. Aguiar & M. J. Feio, 2021. Influence of river regulation and instream habitat on invertebrate assemblage’ structure and function. Science of the Total Environment 794: 148696. https://doi.org/10.1016/j.scitotenv.2021.148696.
Callahan, B. J., P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson & S. P. Holmes, 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13: 581. https://doi.org/10.1038/nmeth.3869.
Certain, G., O. Skarpaas, J.-W. Bjerke, E. Framstad, M. Lindholm, J.-E. Nilsen, A. Norderhaug, E. Oug, H.-C. Pedersen, A.-K. Schartau, G. I. van der Meeren, I. Aslaksen, S. Engen, P.-A. Garnåsjordet, P. Kvaløy, M. Lillegård, N. G. Yoccoz & S. Nybø, 2011. The Nature Index: A General Framework for Synthesizing Knowledge on the State of Biodiversity. PLoS ONE 6: e18930. https://doi.org/10.1371/journal.pone.0018930.
Davy-Bowker, J., R. T. Clarke, R. K. Johnson, J. Kokes, J. F. Murphy & S. Zahrádková, 2006. A comparison of the European Water Framework Directive physical typology and RIVPACS-type models as alternative methods of establishing reference conditions for benthic macroinvertebrates. Hydrobiologia 566: 91–105. https://doi.org/10.1007/s10750-006-0068-5.
Deiner, K., E. Fronhofer, E. Mächler, J. C. Walser & F. Altermatt, 2016. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nature Communications 7: 12544. https://doi.org/10.1038/ncomms12544.
Dudgeon, D., A. H. Arthington, M. O. Gessner, Z.-I. Kawabata, D. J. Knowler, C. Lèvêque, R. J. Naiman, A.-H. Prieur-Richard, D. Soto, M. L. J. Stiassny & C. A. Sullivan, 2006. Freshwater biodiversity: importance, threats, status and conservation challenges. Biological Reviews 81: 163–182. https://doi.org/10.1017/S1464793105006950.
Dunbar, M., A. Ibbotson, I. Gowing, N. McDonnell, M. Acreman & A. Pinder, 2001. Further validation of PHABSIM for the habitat requirements of salmonid fish, UK: Centre for Ecology and Hydrology. R&D Report W6-036, Bristol, United Kingdom.
Elbrecht, V. & F. Leese, 2015. Can DNA-based ecosystem assessments quantify species abundance? testing primer bias and biomass – sequence relationships with an innovative metabarcoding protocol. PloS One 10: e0130324. https://doi.org/10.1371/journal.pone.0130324.
Elbrecht, V. & F. Leese, 2017. Validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment. Frontiers in Environmental Science 5: 11. https://doi.org/10.3389/fenvs.2017.00011.
Elbrecht, V., T. W. A. Braukmann, N. V. Ivanova, S. W. J. Prosser, M. Hajibabaei, M. Wright, E. V. Zakharov, P. D. N. Hebert & D. Steinke, 2019. Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7: e7745. https://doi.org/10.7717/peerj.7745.
Engblom, E. 1996. Ephemeroptera, Mayflies. In Nilsson, A. N. (ed), Aquatic insects of North Europe: A taxonomic handbook, Stenstrup: 13–53
Commission, European, 2000. Directive 2000/60/EC of the European Parliament and of the Council of 23 October 2000 establishing a framework for community action in the field of water policy. Official Journal of European Communities L 327: 1–72.
European Commission, Directorate-General for Environment, A. Hochkirch, A. Casino, L. Penev, D. Allen, L. Tilley, T. Georgiev, K. Gospodinov & B. Barov, 2022. European red list of insect taxonomists. Publications Office of the European Union. https://doi.org/10.2779/364246
European Communities, 2003. Guidance Document No 4. Identification and designation of heavily modified and artificial water bodies. Produced by Working Group 2.2 – HMWB. Office for Official Publications of the European Communities, Luxembourg, Luxembourg.
European Environment Agency, 2019. The European Environment—State and Outlook 2020: Knowledge for Transition to a Sustainable Europe. European Environment Agency, Copenhagen, Denmark.
Fediajevaite, J., V. Priestley, R. Arnold & V. Savolainen, 2021. Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecology and Evolution 11: 4803–4815. https://doi.org/10.1002/ece3.7382.
Frost, S., A. Huni & W. E. Kershaw, 1971. Evaluation of a kicking technique for sampling stream bottom fauna. Canadian Journal of Zoology 49: 167–173. https://doi.org/10.1139/z71-026.
Gleason, J. E., V. Elbrecht, T. W. A. Braukmann, R. H. Hanner & K. Cottenie, 2021. Assessment of stream macroinvertebrate communities with eDNA is not congruent with tissue-based metabarcoding. Molecular Ecology 30: 3239–3251. https://doi.org/10.1111/mec.15597.
Glöer, P., 2002. Süsswassergastropoden Nord- und Mitteleuropas. Bestimmungsschlussel, Lebenweise, Verbreitung. ConchBooks, Hackenheim, Germany.
Goldberg, C. S., C. R. Turner, K. Deiner, K. E. Klymus, P. F. Thomsen, M. A. Murphy, S. F. Spear, A. McKee, S. J. Oyler-McCance, R. S. Cornman, M. B. Laramie, A. R. Mahon, R. F. Lance, D. S. Pilliod, K. M. Strickler, L. P. Waits, A. K. Fremier, T. Takahara, J. E. Herder & P. Taberlet, 2016. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution 7: 1299–1307. https://doi.org/10.1111/2041-210X.12595.
Graf, W., J. Murphy, J. Dahl, C. Zamora-Munoz & M. J. Lopez-Rodriguez, 2008. Distribution and ecological preferences of European freshwater organisms. Volume 1 - Trichoptera. Pensoft Publishers, Sofia, Bulgaria.
Graf, W., A. W. Lorenz, J. M. Tierno de Figueroa, S. Lücke, M. J. Lopez-Rodriguez & C. Davies, 2009. Distribution and ecological preferences of European freshwater organisms. Volume 2 - Plecoptera. Pensoft Publishers, Sofia, Bulgaria.
Haase, P., D. E. Bowler, N. J. Baker, et al., 2023. The recovery of European freshwater biodiversity has come to a halt. Nature 620: 582–588. https://doi.org/10.1038/s41586-023-06400-1.
Hajibabaei, M., T. M. Porter, C. V. Robinson, D. J. Baird, S. Shokralla & M. T. G. Wright, 2019. Watered-down biodiversity? A comparison of metabarcoding results from DNA extracted from matched water and bulk tissue biomonitoring samples. PLoS ONE 1412: e0225409. https://doi.org/10.1371/journal.pone.0225409.
Hering, D., A. Borja, J. I. Jones, D. Pont, P. Boets, A. Bouchez, K. Bruce, S. Drakare, B. Hänfling, M. Kahlert, F. Leese, K. Meissner, P. Mergen, Y. Reyjol, P. Segurado, A. Vogler & M. Kelly, 2018. Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Research 138: 192–205. https://doi.org/10.1016/j.watres.2018.03.003.
Holmen, M. 1987. The Aquatic Adephaga (Coleoptera) of Fennoscandia and Denmark. Brill, Leiden, the Netherlands.
Illumina, 2013. 16S metagenomic sequencing library preparation protocol: preparing 16S ribosomal RNA gene amplicons for the Illumina MiSeq system. Part no. 15044223 Rev B. Illumina, San Diego, CA: https://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf.
Jones, J. I., C. E. M. Lloyd, J. F. Murphy, A. Arnold, C. P. Duerdoth, A. Hawczak, J. L. Pretty, P. J. Johnes, J. E. Freer, M. W. Stirling, C. Richmond & A. L. Collins, 2023. What do macroinvertebrate indices measure? Stressor-specific stream macroinvertebrate indices can be confounded by other stressors. Freshwater Biology 00: 1–16. https://doi.org/10.1111/fwb.14106.
Kjærstad, G., M. Majaneva, M. Falahati-Anbaran, L. Topstad, A. G. Finstad, J. V. Arnekleiv & T. Ekrem, 2022. DNA metabarcoding of preservative ethanol reveals changes in invertebrate community composition following rotenone treatment. Frontiers in Environmental Science 10: 872739. https://doi.org/10.3389/fenvs.2022.872739.
Leese, F., M. Sander, D. Buchner, V. Elbrecht, P. Haase & V. M. A. Zizka, 2021. Improved freshwater macroinvertebrate detection from environmental DNA through minimized nontarget amplification. Environmental DNA 3: 261–276. https://doi.org/10.1002/edn3.177.
Lillehammer, A. 1988. Stoneflies (Plecoptera) of Fennoscandia and Denmark, 21. Brill/Scandinavian Science Press Ltd, Leiden, the Netherlands.
Macher, J.-N., A. Vivancos, J. J. Piggott, F. C. Centeno, C. D. Matthaei & F. Leese, 2018. Comparison of environmental DNA and bulk-sample metabarcoding using highly degenerate cytochrome c oxidase I primers. Molecular Ecology Resources 18: 1456–1468. https://doi.org/10.1111/1755-0998.12940.
Martin, M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 171: 10–12. https://doi.org/10.14806/ej.17.1.200.
McKee, A. M., S. F. Spear & T. W. Pierson, 2015. The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biological Conservation 183: 70–76. https://doi.org/10.1016/j.biocon.2014.11.031.
Mugnai, F., F. Costantini, A. Chenuil, M. Leduc, J. M. Gutiérrez Ortega & E. Meglécz, 2023. Be positive: customized reference databases and new, local barcodes balance false taxonomic assignments in metabarcoding studies. PeerJ 11: e14616. https://doi.org/10.7717/peerj.14616.
Nilsson, A. N. & M. Holmen, 1995. The Aquatic Adephaga (Coleoptera) of Fennoscandia and Denmark, Volume II. Dytiscidea. Brill, Leiden, the Netherlands.
Norwegian Environment Agency, 2018. Klassifisering av miljøtilstand i vann. Økologisk og kjemisk klassifiseringssystem for kystvann, grunnvann, innsjøer og elver. Direktoratsgruppa for gjennomføringen av vanndirektivet, Norway.
Nøst, T., K. Aagaard, J. V, Arnekleiv, J. W. Jensen, J. I. Koksvik & J. O. Solem, 1986. Vassdragsreguleringer og invertebrater. En oversikt over kunnskapsnivået. Økoforsk utredning nr.1. Økoforsk, Trondheim, Norway.
Oksanen, J., F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O'Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs & H. Wagner, 2019. Vegan: Community Ecology Package. https://CRAN.R-project.org/package=vegan.
Piper, A. M., J. Batovska, N. O. I. Cogan, J. Weiss, J. P. Cunningham, B. C. Rodoni & M. J. Blacket, 2019. Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. Gigascience 8: giz092. https://doi.org/10.1093/gigascience/giz092.
Plafkin, J. L., M. T. Barbour, K. D. Porter, S. K. Gross & R. M. Hughes, 1989. Rapid Assessment Protocols for Use in Streams and Rivers: Benthic Macroinvertebrates and Fish. EPAJ444/4-89-001. U.S. Environmental Protection Agency, Washington, United States of America.
Pont, D., M. Rocle, A. Valentini, R. Civade, P. Jean, A. Maire, N. Roset, M. Schabuss, H. Zornig & T. Dejean, 2018. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Scientific Reports 8: 10361. https://doi.org/10.1038/s41598-018-28424-8.
Porter, T. M. & M. Hajibabaei, 2018. Automated high throughput animal CO1 metabarcode classification. Scientific Reports 8: 4226. https://doi.org/10.1038/s41598-018-22505-4.
Ratnasingham S. & P. D. Hebert, 2007. BOLD: The Barcode of Life Data System http://www.barcodinglife.org. Molecular Ecology Notes 7: 355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
R Core Team, 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Reid, A. J., A. K. Carlson, I. F. Creed, E. J. Eliason, P. A. Gell, P. T. J. Johnson, K. A. Kidd, T. J. MacCormack, J. D. Olden, S. J. Ormerod, J. P. Smol, W. W. Taylor, K. Tockner, J. C. Vermaire, D. Dudgeon & S. J. Cooke, 2019. Emerging threats and persistent conservation challenges for freshwater biodiversity. Biological Reviews 94: 849–873. https://doi.org/10.1111/brv.12480.
Resh, V. H. & D. M. Rosenberg, 1993. Freshwater Biomonitoring and Benthic Macroinvertebrates, Chapman and Hall, New York, USA:
Resh, V. H. & J. D. Unzicker, 1975. Water quality monitoring and aquatic organisms: The importance of species identification. Journal (water Pollution Control Federation) 47: 9–19.
Reynoldson, T. B., R. H. Norris, V. H. Resh, K. E. Day & D. M. Rosenberg, 1997. The reference condition: a comparison of multimetric and multivariate approaches to assess water-quality impairment using benthic macroinvertebrates. Journal of the North American Benthological Society 16: 833–852. https://doi.org/10.2307/1468175.
Rinne, A. & P. Wiberg-Larsen, 2017. Trichoptera Larvae of Finland: Identification Key to the Caddis Larvae of Finland and Nearby Countries, Trificon books, Tampere:
Ruppert, K. M., R. J. Kline & M. S. Rahman, 2019. Past, present, and future perspectives of environmental DNA eDNA metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Global Ecology and Conservation 17: e00547. https://doi.org/10.1016/j.gecco.2019.e00547.
Sæther, O. A., 1979. Chironomid communities as water quality indicators. Holarctic Ecology 2: 65–74.
Schmidt-Kloiber, A. & D., Hering, 2015. www.freshwaterecology.info - an online tool that unifies, standardises and codifies more than 20,000 European freshwater organisms and their ecological preferences. Ecological Indicators 53: 271–282. https://doi.org/10.1016/j.ecolind.2015.02.007.
Stoddard, J. L., D. P. Larsen, C. P. Hawkins, R. K. Johnson & R. H. Norris, 2006. Setting expectations for the ecological condition of streams: the concept of reference condition. Ecological Applications 16: 1267–1276. https://doi.org/10.1890/1051-0761(2006)016[1267:SEFTEC]2.0.CO;2.
Sundt-Hansen, L. E., T. Forseth & A. Harby, 2021. Utvidet miljødesign i demovassdrag Nea. HydroCen rapport 22. Norwegian Research Centre for Hydropower Technology, Trondheim, Norway.
Surber, E. W., 1937. Rainbow trout and bottom fauna production in one mile of stream. Transactions of the American Fisheries Society 66: 193–202. https://doi.org/10.1577/1548-8659193666[193:RTABFP]2.0.CO;2.
Swedish Agency for Marine and Water Management, 2018. Bottenfauna i vattendrag – vägledning för statusklassi-ficering. Havs- och vattenmyndighetens rapport 2018:35.
Tharme, R. E., 1996. Review of international methodologies for the quantification of the instream flow requirements of rivers. Water law review. Final report for policy development for the Department of Water Affairs and Forestry, South Africa, Pretoria. Freshwater Research Unit, University of Cape Town, Cape Town, South Africa.
Thrane, J. E., J. Persson, M. Røst Kile, K. A. Bækkelie, K. M. Myrvold, Ø. A. Garmo, M. Grung, J. L. G. Calidonio, H. de Wit & T. F. Moe, 2020. Overvåking av referanseelver 2019. Basisovervåking i henhold til vannforskriften. Miljødirektoratet rapport M-1660. NIVA, Oslo, Norway.
Turner, C. R., M. A. Barnes, C. C. Y. Xu, S. E. Jones, C. L. Jerde & D. M. Lodge, 2014. Particle size distribution and optimal capture of aqueous microbial eDNA. Methods in Ecology and Evolution 5: 676–684. https://doi.org/10.1111/2041-210X.12206.
Van de Bund, W., 2009. Water Framework Directive intercalibration technical report Part 1: Rivers. European Commission Joint Research Centre, Institute for Environment and Sustainability. https://data.europa.eu/doi/https://doi.org/10.2788/23384Vann-Nett, 2022. https://vann-nett.no/portal/, accessed 31.05.2022.
Vitecek, S., R. K. Johnson & S. Poikane, 2021. Assessing the ecological status of European rivers and lakes using benthic invertebrate communities: a practical catalogue of metrics and methods. Water 13: 346. https://doi.org/10.3390/w13030346.
Wallace, J. B., J. W. Grubaugh & M. R. Whiles, 1996. Biotic indices and stream ecosystem processes: results from an experimental study. Ecological Applications 6: 140–151. https://doi.org/10.2307/2269560.
Wang, Q., G. M. Garrity, J. M. Tiedje & J. R. Cole, 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 7316: 5261–5267. https://doi.org/10.1128/AEM.00062-07.
Weigand, H., A. J. Beermann, F. Čiampor, F. O. Costa, Z. Csabai, S. Duarte, M. F. Geiger, M. Grabowski, F. Rimet, B. Rulik, M. Strand, N. Szucsich, A. M. Weigand, E. Willassen, S. A. Wyler, A. Bouchez, A. Borja, Z. Čiamporová-Zaťovičová, S. Ferreira, K.-D.B. Dijkstra & T. Ekrem, 2019. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: gap-analysis and recommendations for future work. Science of the Total Environment 678: 499–524. https://doi.org/10.1016/j.scitotenv.2019.04.247.
Wright, J. F., 2000. An introduction to RIVPACS. In Wright, J. F., D. W. Sutcliffe & M. T Furse (ed), Assessing the biological quality of fresh waters: RIVPACS and other techniques, Ambleside: 1–24.
Zhang, G. K., F. J. J. Chain, C. L. Abbott & M. E. Cristescu, 2018. Metabarcoding using multiplexed markers increases species detection in complex zooplankton communities. Evolutionary Applications 11: 1901–1914. https://doi.org/10.1111/eva.12694.
Zhang, Z., S. Schwartz, L. Wagner & W. Miller, 2000. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology 7: 203–214. https://doi.org/10.1089/10665270050081478.
Zizka, V. M. A., F. Leese, B. Peinert & M. F. Geiger, 2018. DNA metabarcoding from sample fixative as a quick and voucher-preserving biodiversity assessment method. Genome 62: 122–136. https://doi.org/10.1139/gen-2018-0048.
Acknowledgements
This study received financial support from the Norwegian Research Centre for Hydropower Technology (HydroCen), Research Council of Norway project number 257588. We are grateful to anonymous reviewers who contributed significantly to improve the manuscript.
Funding
Open access funding provided by Norwegian institute for nature research.
Author information
Authors and Affiliations
Contributions
FF, TB, and LESH conceived the ideas and designed the methodology; TB and RS collected the data; TB and MM analysed the data; MM took the lead in writing the manuscript with contributions and revisions of all authors. All authors gave their final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Handling editor: Verónica Ferreira
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Majaneva, M., Sundt-Hansen, L.E., Brandsegg, H. et al. Comparing methods and indices for biodiversity and status assessment in a hydropower-regulated river. Hydrobiologia 851, 2139–2157 (2024). https://doi.org/10.1007/s10750-023-05448-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05448-4