Abstract
The basal splits within Syndermata probably date back hundreds of millions of years, and extant syndermatans greatly differ in reproduction modes (parthenogenesis, metagenesis, heterosexuality) and lifestyles (free-living, epizoic, endoparasitic). Against this background, the present investigation aims to reconstruct genome and transcriptome metrics in the last common ancestor (LCA) of Syndermata. We especially annotated nuclear genome assemblies of one representative, each, of monogononts (Brachionus plicatilis), bdelloids (Adineta vaga), seisonids (Seison nebaliae), and acanthocephalans (Pomphorhynchus laevis), S1 and additionally considered previously published data. Present results suggest that the syndermatan LCA possessed a compact nuclear genome of 50–100 Mb with a coding portion of ≥ 40%. Its ~ 17,000 protein-coding genes probably had about 4–6 exons and 3–5 introns on average. Long interspersed nuclear elements (LINEs) should have occupied 4–5% of the ancestral genome. Based on this, independent evolutionary trajectories involving gene loss, genome duplication, and repeat proliferation resulted in greatly varying genomes within the clade. Nevertheless, we identified 2114 clusters containing proteins of all four syndermatan taxa. About 200 of these clusters lacked sufficiently similar counterparts in the platyhelminths Macrostomum lignano and Schmidtea mediterranea. Gene ontologies assigned to these clusters might reference the muscular pharynx, light perception, and transposition, amongst others.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rotifera sensu lato or Syndermata is a monophyletic, albeit very diverse, metazoan clade (Ahlrichs, 1997; Herlyn & Ehlers, 1997; Sørensen & Giribet, 2006; Wey-Fabrizius et al., 2014). While competing hypotheses on the internal phylogeny exist, four monophyletic syndermatan groups are commonly recognized: Monogononta, Bdelloidea, Seisonidea, and Acanthocephala (von Haffner, 1950; Rieger & Tyler, 1995; Ahlrichs, 1997; Herlyn et al., 2003; Sørensen & Giribet, 2006; Fontaneto & De Smet, 2015). Most probably, the last common ancestor of Syndermata had a syncytial epidermis, just as it is the case in all extant members of the clade. In addition, the syndermatan LCA should have possessed a muscular pharynx with cuticular jaw elements (mastax) (e.g., Fontaneto & De Smet, 2015; Herlyn, 2021). Still, the basal splits within the clade probably date back hundreds of millions of years (Herlyn, 2021), and the extant species greatly differ in lifestyle, ecology, and modes of reproduction. Species of Acanthocephala (thorny-headed worms) are obligate endoparasites with complex lifecycles involving Mandibulata (Arthropoda) as intermediate and Gnathostomata (Vertebrata) as definitive hosts. The endoparasitic lifestyle is reflected in morphological peculiarities such as a so-called proboscis at the anterior body pole (Taraschewski, 2015; Herlyn, 2021), with which the adults attach to the digestive tract wall of their host (Hammond, 1966; Herlyn et al., 2001; Herlyn & Taraschewski, 2017). In addition, the few extant species of Seisonidea (also Seisonacea, Seisonidae) live epizoically, potentially also ectoparasitically on marine crustaceans (Leptostraca) of the genus Nebalia (Leasi et al., 2012; Ahlrichs & Riemann, 2018). Acanthocephalans and seisonids share a strictly heterosexual mode of reproduction and consistently lack a wheel organ or corona, which is present in the other two syndermatan taxa, Bdelloidea and Monogononta. Bdelloid and monogonont species are predominantly free-living and occur mainly, but not exclusively, in freshwater (Fontaneto & De Smet, 2015). However, there are also significant differences between monogononts and bdelloids. For example, no males have been observed in bdelloids, which appear to reproduce exclusively by parthenogenesis (Wallace, 2002; Mark Welch & Meselson, 2003; Mark Welch et al., 2004; Flot et al., 2013). Yet, monogononts can switch from parthenogenesis to sexual reproduction upon environmental deterioration (Wallace, 2002; Fontaneto & De Smet, 2015; Franch-Gras et al., 2019). Along with these differences, most molecular studies indicate that Monogononta and Bdelloidea do not share a monophyletic origin, thus rejecting monophyletic Eurotatoria (Monogononta + Bdelloidea) (Mark Welch, 2000; Witek et al., 2008). In contrast, the phylogenetic position of Seisonidea inside Syndermata was not stable in molecular analyses, and alternated between a basal branching (e.g., Struck et al., 2014) and a sister-group relationship with Acanthocephala (Herlyn et al., 2003; Sørensen & Giribet, 2006; Wey-Fabrizius et al., 2014; Sielaff et al., 2016) within so-called Pararotatoria (Zrzavý, 2001).
Until recently, about one hundred nuclear genomes of free-living monogononts and bdelloids have been reconstructed from next generation sequencing data (2023-15-02 https://www.ncbi.nlm.nih.gov/assembly). The number of assemblies is continuously increasing, thus reflecting considerable attention paid to this group of animals. Interest has attracted that monogononts show great variability in genome size, with haploid genomes ranging from ~ 51 to ~ 115 Mb in the Brachionus plicatilis Müller, 1786 complex (Blommaert et al., 2019). Furthermore, abovementioned parthenogenesis in bdelloids has attracted much attention (Wallace, 2002; Mark Welch & Meselson, 2003; Franch-Gras et al., 2019), as has the degenerate state of tetraploidy in this taxon (Mark Welch et al., 2008; Hur et al., 2009; Flot et al., 2013; Nowell et al., 2018; Han et al., 2019). However, most studies have focused on monogonont and bdelloid genomes and transcriptomes, whereas comprehensive data on acanthocephalans and seisonids have only recently been added. The first nuclear genome assembly of an acanthocephalan was found to be surprisingly large for a parasite (~ 260 Mb), thereby displaying a high share of repetitive stretches (Mauer et al., 2020). On the contrary, the first nuclear genome assembly of a seisonid (~ 44 Mb) belonged to the smallest within Syndermata (Mauer et al., 2021). Previous genome annotations further illustrated strong differences in gene number depending on the bioinformatic pipeline used. For example, 49,300 (Flot et al., 2013) and 67,364 (Nowell et al., 2018) genes were annotated for the bdelloid Adineta vaga (Davis, 1873). Likewise, for B. plicatilis, 54,725 (Franch-Gras et al., 2018), 20,154 (Han et al., 2019), and 12,484 (Blommaert et al., 2019) genes were annotated, although the genome assemblies analyzed had similar sizes, with a range from 99 to 108 Mb.
Our aim here is to comparatively analyze the nuclear genome structure and content within Syndermata. Accounting for alternative views on the detailed phylogenetic relationships inside the clade, our focus is on states in the last common ancestor (LCA) of Syndermata. To achieve this goal, we applied the same annotation pipeline on nuclear genome assemblies of the endoparasitic acanthocephalan Pomphorhynchus laevis (Zoega in Müller, 1776) Porta 1908, the epizoic seisonid Seison nebaliae Grube, 1861, and two free-living wheel animal species, the monogonont B. plicatilis and the bdelloid A. vaga. Published data on genome size and organization as well as gene and repeat repertoires are considered too. This includes additional species of Protostomia such as the free-living flatworms Macrostomum lignano Ladurner, Schorer, Salvenmoser & Rieger, 2005 and Schmidtea mediterranea Benazzi, Baguna, Ballester & del Papa, 1975 (Platyhelminthes). Furthermore, we delineate independent evolutionary trajectories inside Syndermata, thus shedding light on the evolution history of this very diverse clade.
Materials and methods
Syndermatan genome assemblies and annotation of their repetitive portions
We analyzed one genome assembly for each of the main syndermatan taxa. If multiple assemblies were available, we chose the most contiguous one, thus laying the grounds for an as complete annotation as possible (Yandell & Ence, 2012). The approach resulted in the inclusion of nuclear genome assemblies of P. laevis (sensu lato in accordance with Perrot-Minnot et al., 2019) as representative for Acanthocephala (Mauer et al., 2020), S. nebaliae for Seisonidea (Mauer et al., 2021), B. plicatilis for Monogononta (Han et al., 2019), and A. vaga for Bdelloidea (Simion et al., 2021) (for details, see Table 1). The genome assemblies of P. laevis and S. nebaliae underwent additional filtering before annotation (Supplementary Material 5). Repetitive regions (transposable elements and multiple simple repeats) were soft-masked in the B. plicatilis and A. vaga nuclear genomes according to respective models for P. laevis (Mauer et al., 2020) and S. nebaliae (Mauer et al., 2021), using RepeatMasker Open v4.0.7 (https://usegalaxy.org/; Smit & Hubley, 2013). The procedure included the generation of separate repeat databases for single species with dnaPipeTE (Goubert et al., 2015) and RepeatModeler v2.0.1 (Smit & Hubley, 2008) (see Supplementary Material 5 for details).
Mapping of RNA-seq reads
For the four syndermatan taxa focused, we used transcriptome in addition to nuclear genome assemblies as specified in Table 1. In the case of A. vaga, we considered three assemblies representing different desiccation stages (Hecox-Lea & Mark Welch, 2018), to include a comprehensive representation of transcribed loci. Likewise, several specimens were represented in the transcriptomes of the other three taxa considered. Quality control of RNA-seq reads was performed with Fastqc v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We removed overrepresented mitochondrial sequences using bbduk v38.73 (k = 31 hdist = 1; https://sourceforge.net/projects/bbmap/). Trimming was done with Trimmomatic v0.39 (Bolger et al., 2014), using the following parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 AVGQUAL:20 MINLEN:40. Filtered and trimmed RNA reads were aligned in two-pass mode to the respective soft-masked genome assembly using the program STAR v2.7.6a (Dobin et al., 2013).
Annotation of syndermatan genomes
For preliminary annotations, we used BRAKER1 v2.1.6 (Stanke et al., 2006, 2008; Li et al., 2009; Barnett et al., 2011; Buchfink et al., 2014; Lomsadze et al., 2014; Hoff et al., 2016, 2019), BRAKER2 (Brůna et al., 2021), and TSEBRA (Gabriel et al., 2021). Eventually, we relied on BRAKER1 since the resulting metrics as obtained with the AGAT toolkit v0.7.0 (agat_sp_statistics.pl; https://github.com/NBISweden/AGAT) came close to values in previous publications (for details, see Discussion). Corresponding annotations also appeared superior regarding numbers of recovered BUSCO genes (not shown). We determined the number of bases marked as repetitive (soft-masked) for coding sequences (CDSs), intronic regions, and intergenic regions. Furthermore, translated transcripts were matched with the SwissProt database via BLASTP to identify hits in other organisms (downloaded on 2021-07-12; E value ≤ 1e−03). We only considered the longest annotated transcript of a gene, thus avoiding multiple characterization of genomic regions due to different isoforms. Annotations for P. laevis and S. nebaliae were manually curated and genes with extreme repeats and isoforms lacking a start or stop codon were removed. We additionally checked if comparatively long intron sequences in the nuclear genome assembly of P. leavis might contain open reading frames (ORFs) of unannotated proteins. Corresponding sequences were matched via BLASTX to the SwissProt database (as of 2021-07-12, E value ≤ 1e−03). Annotation files can be downloaded from the journal website as Supplementary Materials 1 for the monogonont, 2 for the bdelloid, 3 for the seisonid, and 4 for the acanthocephalan.
Clustering of orthlogues and paralogues
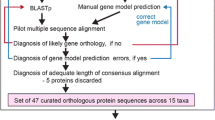
Translated transcript datasets were used to cluster putative orthologues and paralogues with the OrthoVenn2 (Xu et al., 2019) web server (utilized 01/20-23: https://orthovenn2.bioinfotoolkits.net/home). OrthoVenn2 conducts an all-against-all-protein sequence comparison with DIAMOND v0.9.24 (Buchfink et al., 2014) and clusters orthologues and paralogues (orthogroups) based on sequence similarity (Wang et al., 2015; Xu et al., 2019). The web server was started with default values (E value ≤ 1e−02, inflation value = 1.5). The resulting clusters contained sequences of a single up to all four species examined. Single-species clusters can be taken as paralogues, while multi-species transcript clusters contain orthologues but may also contain paralogues. First, we performed clustering with the datasets of the four syndermatan species focused, to identify, amongst others, the clusters which were likely present in their LCA. Second, we ran OrthoVenn2 on translated transcript lists of the longest isoforms of the four syndermatan species and on translated transcriptomes (PRJNA284736, PRJNA885486) of two free-living platyhelminths, M. lignano (Platyhelminthes, Macrostomidae) and S. mediterranea (Platyhelminthes, Dugesiidae). The choice here was since Platyhelminths belong to the closest phylogenetic relatives of Syndermata (e.g., Struck et al., 2014), for which the transcriptomes have been extensively studied. Addition of these outgroup representatives was intended to harness protein clusters in the syndermatan LCA the genes of which might have emerged since the split from the platyhelminth lineage. We also used OrthoVenn2 for allotting gene ontologies (GOs) to single protein clusters via SwissProt hits.
Syndermatan genome metrics within the larger Protostomia context
In the Discussion, we compare single genome metrics between Syndermata and other Protostomia. For this purpose, we gathered genome sizes and gene numbers of the abovementioned flatworm species M. lignano (ML2, dv1 strain) and S. mediterranea (schMedS3_haplotype1, S2F19 strain) from WormBase ParaSite (accessed on 2023-04-18). We additionally considered corresponding data for the free-living roundworm Caenorhabditis elegans (Maupas, 1900) Dougherty, 1953 (Edysozoa, Nematoda, Rhabditina) and the insect Drosophila melanogaster Meigen, 1830 (Exdysozoa, Hexapoda, Diptera) as reported at Ensembl genome browser 109 (WBcel235, BDGP6.32). Data on additional species were extracted from the literature cited.
Results
Syndermatan genome assemblies and repetitive portions
The nuclear genome assemblies of the syndermatan representatives included in this study differed considerably in size, but also in respect to the repetitive portions (Tables 1, 2). With about 253 Mb, the genome assembly of the acanthocephalan (P. laevis) was the largest, followed by the ones of the monogonont (B. plicatilis) with ca. 107 Mb and the bdelloid (A. vaga) with ca. 101 Mb. With about 44 Mb, the genome assembly was smallest in the seisonid (S. nebaliae). The genome assemblies of A. vaga and S. nebaliae consistently had larger CDS and smaller intronic proportions, whereas the ratio was inverse in the other two syndermatan genome assemblies. Furthermore, the total CDS proportion amounted to 45% in the S. nebaliae genome assembly, while the fraction dropped to 5% in the genome assembly for P. laevis. With CDS percentages of 22% and 41%, the genome assemblies representing both free-living species, B. plicatilis and A. vaga, ranged in-between. In addition, 42% to 63% of the genomes did not receive any annotation by the pipeline applied and hence were classified as intergenic. Corresponding proportions were most similar in S. nebaliae and A. vaga, slightly increased in B. plicatilis, and reached a maximum value in P. laevis (Fig. 1; Table 2).
Spine plot illustrating the organization of nuclear genome assemblies of four syndermatan species. The y-axis shows cumulative percentages. The width of the bars is proportional to absolute numbers. Color code refers to coding sequences (CDS), intronic (intr.), and intergenic sections (interg.). Genome assemblies of the bdelloid (A. vaga) and the seisonid (S. nebaliae) consistently showed larger CDS than intronic shares, while the inverse relation held for the monogonont (B. plicatilis) and the acanthocephalan (P. laevis). The plot was generated with ggplot2 (Wickham, 2016) utilizing scales (Wickham & Seidel, 2022) within the R environment (R Core Team, 2022)
As to be expected, the genome assemblies were to a higher proportion repetitive in non-coding (intergenic and intronic) than coding regions. Moreover, long interspersed nuclear elements (LINEs) formed the largest fraction of classified transposable elements in all four species. Nevertheless, LINEs made up minor fractions of 4–5% of the genome assemblies in A. vaga, B. plicatilis, and S. nebaliae. With 37%, LINEs occupied a clearly larger proportion of the genome assembly of the acanthocephalan P. laevis (Fig. 2; Supplementary Material 5). Correspondingly, the greater size of the P. laevis genome assembly was essentially due to its disproportionately large repetitive fraction (Table 2). In fact, the non-repetitive portion varied by a factor of 2.4 only, whereby the span was ~ 37–89 Mb (Table 1). Overall, repeat annotations were most similar between the nuclear genome assemblies of A. vaga and S. nebaliae, despite a roughly two-fold difference in size (Fig. 2).
Donut plot of repetitive and non-repetitive portions in the nuclear genome assemblies of four syndermatan species. Each cycle shows one assembly. Green and blue tones correspond to repetitive portions, while yellow highlights non-repetitive shares. Shares of individual repeat classes were most similar in the nuclear genome assemblies of the bdelloid (A. vaga) and seisonid (S. nebaliae). Also, the entire repetitive portions were smallest in these two assemblies, followed by the one for the monogonont (B. plicatilis) and the acanthocephalan (P. laevis). The plot was created with ggplot2 (Wickham, 2016) within the R environment (Team, 2022). The data depicted were based on annotations with RepeatMasker Open-4.0.7 (https://usegalaxy.org/; Smit & Hubley, 2013). For details, see Supplementary Material 5. LINEs, long interspersed nuclear elements; LTRs, long terminal repeats
Annotation of protein-coding genes
More than 80% of the RNA-seq reads were mapped to the respective nuclear genome assemblies for B. plicatilis, A. vaga, and P. laevis. In S. nebaliae, this percentage was not half as high (Table 1), which presumably reflected the representation of two strains in the RNA reads mapped (for details, see Mauer et al., 2021). In the four assemblies, nearly all splice sites showed canonical start and end motifs (GT and AG). Moreover, at least half of the predicted protein sequences had matches in the SwissProt database, as revealed by BLASTP search (Table 2). Total numbers of predicted protein-coding genes were lowest in S. nebaliae (N = 11,502) and P. laevis (12,073), intermediary in B. plicatilis (16,800), and highest in A. vaga (29,732). Average per transcript numbers of exons (4.3–6.1) and introns (3.4–5.1) were in similar ranges in the four syndermatan genome assemblies (Table 2).
Frequency distributions in A. vaga were entirely (exon length), almost completely (gene and CDS lengths), or at least partially (intron length) above the corresponding curves in the other three taxa. In terms of the shape, graphs for the parasite P. laevis were most dissimilar from the ones for the other three species (Fig. 3). For example, the intron size distribution in P. laevis tended to be bimodal, with a maximum at small sizes and a second flat shoulder in the range of higher values. In contrast, the intron size distributions of the other three syndermatan species appeared monomodal, with a maximum in the region of small sizes. The general dissimilarity of the distributions in the acanthocephalan on the one hand and the other three taxa on the other hand was also evident in single metrics. Thus, exons were overall shorter in P. laevis (median: 120 bp) than in the other three taxa (141–170 bp). Conversely, introns were overall longer in the acanthocephalan (median: 868 bp) compared to the monogonont, bdelloid, and seisonid (50–58 bp). Correspondingly, the acanthocephalan genome assembly displayed the longest genes (median: 2537 bp) while CDS size was shortest (699 bp), overall. Respective medians ranged from 1370 to 1479 bp (gene length) and 1047 to 1200 bp (CDS length) in the other three species (Table 2).
Frequency distributions of A exon, B intron, C gene, and D CDS lengths in the nuclear genome assemblies of four syndermatan species. Ordinate values correspond to non-overlapping intervals of 100 bp. In detail, 1 represents 1–100 bp, 2 represents 101–200 bp, 3 represents 201–300 bp, etc. Graphs are truncated to the right. Two general trends emerge: Firstly, curves for the bdelloid (A. vaga) tend to be above the ones in the monogonont (B. plicatilis), seisonid (S. nebaliae, S. neb.), and acanthocephalan (P. laevis). Secondly, the shape of the curves is most dissimilar in P. laevis, relative to their counterparts in the other three species. Annotations had been carried out with BRAKER1
Despite non-normality of the distributions (Fig. 3), the mean values confirmed the special status of P. laevis as exemplified by a higher average intron length in the acanthocephalan (1817 bp) than the other three species (120–504 bp). Particularly long introns in P. laevis prompted us to screen for potentially hidden CDS by BLASTX against the SwissProt database. Applying an E value threshold ≤ 0.001, this search revealed matches for only about 8% of the intronic sequences (not shown). Closer examination uncovered that the vast majority (82%) of the matched query sequences were soft-masked to at least half of the bases, and that most of the matching sequences in the SwissProt database related to transposons.
Syndermatan protein clusters
Based on the longest transcript of each gene, we inferred orthogroups, i.e., clusters of orthologous and/or paralogous genes (Fig. 4A). The program applied, OrthoVenn2, identified a total of 11,869 orthogroups, thereof 6290 containing sequences of two or more species. In confirmation of the suitability of the pipeline, the intersection of all four syndermatan taxa contained the highest number of clusters (N = 2114). On average, these core orthogroups contained 1.08, 1.21, and 1.33 sequences of S. nebaliae, P. laevis, and B. plicatilis, respectively. With 1.77, the corresponding number was not quite twice as high in A. vaga (Supplementary Material 5). It should be mentioned that the intersection of A. vaga, S. nebaliae, and P. laevis and consequently of Bdelloidea, Seisonidea, and Acanthocephala (Hemirotifera) contained 132 protein clusters, and that of S. nebaliae and P. laevis or consequently of Seisonidea and Acanthocephala (Pararotatoria) 41. Moreover, OrthoVenn2 derived the highest number of paralogue clusters for the bdelloid (Fig. 4A), which also had the highest total number of clusters (Fig. 4B). Disregarding this special pattern in the degenerate tetraploid there was a general trend for reduced cluster numbers from B. plicatilis via S. nebaliae to P. laevis. Most orthogroups contained transcripts of B. plicatilis (91%) and A. vaga (94%), while S. nebaliae was represented in considerably less (66%), and P. laevis in about half of the clusters (Supplementary Material 5). Nevertheless, the fraction of genes in core orthogroups relative to the total number of predicted genes was slightly higher in P. laevis and S. nebaliae than B. plicatilis and A. vaga (Fig. 5).
Clustering of proteins of four syndermatan representatives. A Venn diagram of clustered proteins. The areas are semi-proportional to the number of orthogroups. The intersection containing clusters with sequences of all four syndermatan species considered, i.e., the monogonont (B. plicatilis), the bdelloid (A. vaga), the seisonid (S. nebaliae), and the acanthocephalan (P. laevis), was the largest. Furthermore, A. vaga had the most private clusters. B Bar chart of total numbers of clusters per species. Again, A. vaga stood out, by having the highest total number of clusters. The other bars reflected a trend for decreasing total cluster numbers from the free-living monogonont (B. plicatilis) via the seisonid living on crustaceans (S. nebaliae) to the endoparasitic acanthocephalan (P. laevis). Clustering analysis was conducted with OrthoVenn2. The semi-proportional Venn diagram was drawn with nVennR (Pérez-Silva et al., 2018) and rsvg (Ooms, 2022) within the R environment (R Core Team, 2022)
Spine plot visualizing percentages of clustered and unclustered proteins in four syndermatan taxa. The y-axis refers to cumulative percentages. The width of the bars corresponds to absolute numbers of genes. Color code gives core orthogroups containing proteins of all four species (core), orthogroups shared between two or three species (2–3), one-species clusters with presumed paralogues (p), and unclustered sequences (uc). Genome assemblies of the seisonid (S. nebaliae) and the acanthocephalan (P. laevis) displayed highest percentages of transcripts within core orthogroups, followed by the monogonont (B. plicatilis). The plot was created with ggplot2 (Wickham, 2016) utilizing scales (Wickham & Seidel, 2022) within the R environment (R Core Team, 2022)
Subsequent OrthoVenn2 analysis integrating the datasets of M. lignano and S. mediterranea revealed 194 clusters with monogonont, bdelloid, seisonid, and acanthocephalan proteins for which no sufficiently similar platyhelminth counterparts were identified. The corresponding GOs contained 4–34 syndermatan proteins, each. The GOs of the twenty largest of these clusters repeatedly referenced the muscular system (positive regulation of sarcomere organization, regulation of muscle contraction, striated muscle cell development) and food intake (suckling behavior). Light perception was another recurrent entanglement of the top-abundant protein clusters, as well as, post-translational modification, splicing, transposition, cell proliferation, sexual reproduction, nervous system development, development nutrition, etc. (Table 3). Additional GOs referenced signaling, ion transport, catalysis, etc. (Supplementary Material 5).
Discussion
Reproducibility of genome annotations
Using the same pipeline, we comparatively analyzed nuclear genome assemblies of the free-living wheel animals B. plicatilis (Monogononta) and A. vaga (Bdelloidea), the epizoon S. nebaliae (Seisonidea), and the endoparasite P. laevis (Acanthocephala). The genome metrics derived were widely consistent with previous estimates. For example, current and previous estimates on total CDS size in B. plicatilis were close to each other (Han et al., 2019: 26.1 Mb; this study: 23.9 Mb). In addition, the number of genes predicted here for A. vaga (29,732) was close to the one published before (Simion et al., 2021: 31,582). Also, proportions of repetitive (soft-masked) regions as derived by us were similar to corresponding values in preceding studies. For instance, repetitive portions were given with 16% (Kim et al., 2018), 20% (Nowell et al., 2021), and 16% (this study) for A. vaga, 32% (Kang et al., 2020) and 30% (this study) for B. plicatilis, and 66% (Nowell et al., 2021) and 65% (this study) for P. laevis. Not least, our data reflect a trend towards larger introns and intergenic regions in greater genomes (Tables 1, 2) which was already demonstrated for diverse representatives of Eukaryota (Francis & Wörheide, 2017) including monogononts (Blommaert et al., 2019). We take these consistencies as an affirmation that the pipeline applied yielded representative results. In fact, the close match of the current and previously published genome metrics let us rely on BRAKER1 annotations, after having applied additional pipelines (BRAKER2, TSEBRA) in preliminary analyses (Gabriel et al., 2021; Brůna et al., 2021). BRAKER1 annotations also appeared superior considering numbers of BUSCO gene completeness (not shown).
Size and structure of syndermatan genomes
Syndermatan nuclear genomes display great variability in size across and within the four clades addressed here, i.e., Monogononta, Bdelloidea, Seisonidea, and Acanthocephala. For example, genome size is known to vary by a factor of up to seven within monogononts of the B. plicatilis species complex (Blommaert, 2020). Likewise, genome sizes of ~ 96 to ~ 1193 Mb, corresponding to a 12-fold difference, have been reported for bdelloids (Pagani et al., 1993; Mark Welch & Meselson, 1998, 2003; Dolezel et al., 2003; Simion et al., 2021). Furthermore, very small nuclear genomes were known before to occur in syndermatans as exemplified by 32.7 Mb in the monogonont Proales similis de Beauchamp, 1907 (Kim et al., 2017). Thus, genome size divergence as observed here (~ 44–253 Mb) is not unusual in Syndermata. Also, there appears to be a general tendency for a compact organization of the nuclear genomes in syndermatans. For example, the 10,785 annotated protein-coding genes in P. similis were found to occupy about 18 Mb or 55% of the genome assembly (Kim et al., 2021). Compact genomes with CDS proportions of about 40% actually seem to be quite common in monogononts and bdelloids (Nowell et al., 2018; Byeon et al., 2021; Kim et al., 2021). Correspondingly, the present investigation revealed CDS proportions of 41% and 45% in the bdelloid A. vaga and the seisonid S. nebaliae, respectively (Table 2). The syndermatan LCA might thus have possessed a likewise compact nuclear genome, with a CDS portion of 40% or above.
Notably, haploid genome size in A. vaga is just over 100 Mb (Simion et al., 2021; present study). Yet, this size already reflects genome duplication in the bdelloid stem line (Mark Welch et al., 2008; Hur et al., 2009; Flot et al., 2013; Nowell et al., 2018; Simion et al., 2021). The starting point prior to duplication might hence have been a genome of about 50 Mb, which would be in the range of the smallest syndermatan genome assemblies published to date (see above). In the B. plicatilis species complex, larger nuclear genomes would then have evolved by satellite DNA proliferation (Blommaert et al., 2019; Stelzer et al., 2021a). Likewise, the comparably large nuclear genome assembly of the acanthocephalan P. laevis (ca. 253 Mb) seems to reflect a significant expansion of the repetitive portion (65% soft-masked). The latter might appear unexpected, as parasite evolution is usually believed to be accompanied by genome size reduction (Katinka et al., 2001; Kikuchi et al., 2011; Poulin & Randhawa, 2015). However, a pattern such as in P. laevis is not unique (Spanu et al., 2010). In any case, the adaptive gain of genome size expansion inside Syndermata remains elusive, albeit some correlations might bear hints. For example, sessile species were reported to tentatively have larger genomes than free-living ones. In addition, colonial species were found to have larger genomes than their solitary counterparts (Brown & Walsh, 2019). Furthermore, individual size, egg size, and embryonic developmental time might positively correlate with genome size (Stelzer et al., 2021b). Thus, genome size evolution in syndermatans could reflect changes in lifestyle and life history traits.
Considering several intermediary genome metrics for B. plicatilis (Tables 1, 2), genome size in the syndermatan LCA might alternatively have been in the range of this species (~ 100 Mb). Even under this premise, the ancestral syndermatan genome would still have been much smaller than the genomes of extant free-living platyhelminths like M. lignano (WormBase ParaSite: 1,040,124,789 bp; Wasik et al., 2015) and S. mediterranea (WormBase ParaSite: 839,952,239 bp; Grohme et al., 2018). Nevertheless, much smaller genomes occur in parasitic platyhelminths as exemplified by monogenean flukes (Monogenea) such as Gyrodactylus salaris Malmberg, 1957 (Hahn et al., 2014: ~ 120 Mb) and tapeworms (Cestoda) of the genus Taenia Linnaeus, 1758 (Wang et al., 2016: ~ 170 Mb). Moreover, a genome of ~ 100 Mb in the syndermatan LCA would approximate the corresponding sizes in D. melanogaster (Ensembl 109: 143,726,002 bp; Adams et al., 2000) and in the free-living nematode C. elegans (Ensembl 109: 100,286,401 bp; Waterston & Sulston, 1995). Accounting for both options discussed here, the LCA of Syndermata might have had a nuclear genome of 50–100 Mb.
Syndermatan genes
The present finding of the highest number of nearly 30,000 protein-coding genes in the A. vaga genome is in accordance with a degenerate tetraploid state of this bdelloid (Mark Welch et al., 2008; Hur et al., 2009; Flot et al., 2013; Nowell et al., 2018; Vakhrusheva et al., 2020; Simion et al., 2021). Indeed, clearly fewer genes (11,502–16,800) were predicted for the other syndermatan genome assemblies (Table 2). Bdelloid tetraploidy was further reflected in overall higher frequency distribution curves for exon, intron, gene, and CDS lengths (Fig. 3). In addition, we annotated approximately 500 more genes in the endoparasite P. laevis than in S. nebaliae. This might hint to stronger ties in the latter to the crustacean than commonly assumed, and an ectoparasitic rather than epizooic lifestyle (Illgen, 1916; Segers & Melone, 1998; Ahlrichs & Riemann, 2018). However, there might be another explanation considering the prominent share of LINEs in the P. laevis genome assembly. Thus, new genes might have emerged on the lineage to P. laevis through LINE1 mediated exon shuffling (Gilbert, 1978; Moran et al., 1999; Patthy, 1999). Considering the specialties in bdelloid, seisonid, and acanthocephalan evolution, gene number in the monogonont B. plicatilis (Table 2: 16,800) might be closest to the ancestral syndermatan state. Accounting for potential non-representation of some genes in the monogonont assembly and non-detection of additional ones, the syndermatan LCA might have possessed about 17,000 protein-coding genes. These might have had 4–6 exons and 3–5 introns on average as suggested by widely congruent results across the four genomes assemblies analyzed here (Table 2).
Gene numbers similar in magnitude to the present estimate for the syndermatan LCA have been reported for other helminth taxa. For example, about 15,000 homologous genes were identified in Rhabditina, Tylenchina, Spirurina, and Dorylaimia (Ecdysozoa, Nematoda) (Parkinson et al., 2004). However, 28 of the 30 roundworm species included in the latter study were parasites. The annotation of almost 20,000 protein-coding genes in the free-living nematode C. elegans (Rhabditina) might thus be closer to the ancestral nematode state (Ensembl 109). In line with this, around 20,000 coding genes were annotated in the genome of another ecdysozoan representative, the beetle Tenebrio molitor Linnaeus, 1758 (Coleoptera) (Kaur et al., 2022). Still, this does not necessarily correspond to the ancestral gene number of hexapods as illustrated by ~ 14,000 genes in the fly D. melanogaster (Ensembl 109). Furthermore, higher numbers of coding genes have been reported for closer phylogenetic relatives of Syndermata, flatworms (Platyhelmintes) (e.g., Struck et al., 2014). For example, single species of abovementioned genus Macrostomum Schmidt, 1848 might have ~ 24,000 homologous genes (Brand et al., 2020; also, Wasik et al., 2015), and almost 21,400 coding genes were annotated in the genome of the platyhelminth S. mediterranea (WormBase ParaSite; Grohme et al., 2018). These counts demonstrate that ~ 17,000 genes as suggested here for the syndermatan LCA would be within the expectable range for a protostome. If about 20,000 coding genes represent the ancestral state, about 3000 might have gone lost on the lineage to the syndermatan LCA since the split from the lineage to platyhelminths.
Syndermatan protein clusters
If no outgroup species were included, OrthoVenn2 inferred the largest number of clusters for the intersection between the four syndermatans (Fig. 4A). Obviously, these core orthogroups (N = 2114) refer to proteins with sufficiently conserved sequences to be clustered. The same should apply to the smaller numbers of protein clusters representing the intersections of A. vaga, S. nebaliae, and P. laevis and S. nebaliae and P. laevis. Part of the corresponding genes might also have emerged in the stem lines of Syndermata, Hemirotifera (Bdelloidea + Seisonidea + Acanthocephala) and Pararotatoria (Seisonidea + Acanthocephala). However, higher sequence divergence and incompleteness of the transcriptome datasets might also have hindered clustering across larger species combinations. This might especially have affected the second OrthoVenn2 run which additionally included the transcriptomes of two free-living platyhelminths. Either way, the addition of both outgroup representatives helped us to differentiate between conserved orthogroups (including sequences from all syndermatan representatives plus outgroup representatives) and orthogroups which might have emerged on the lineage to Syndermata or Gnathifera (including sequences from all syndermatan representatives but not the outgroup species). Alternatively, the corresponding genes might have experienced stronger divergence after the split of the lineages to Syndermata and Plathyhelminthes. The respective GOs might hold clues to the biology of their LCA (Table 3; Supplementary Material 5). Thus, gene ontologies referencing striated musculature and sarcomere development could hint to the muscular pharynx or mastax bearing jaw-like solid parts. These probably emerged in the stem line of Gnathifera and should have been retained in the syndermatan LCA (Ahlrichs, 1997; Herlyn & Ehlers, 1997; Fontaneto & De Smet, 2015; Sørensen et al., 2016). The GO of the largest of these orthogroups, suckling behavior, should be taken with caution, but could still be another indication of food intake. Moreover, GOs referencing light perception appear plausible in respect to the probable abilities of the LCA of Syndermata (Clément & Wurdak, 1991; Fontaneto & De Smet, 2015). That syndermatan genomes contain transposons and thus testify to ample transposition events was also shown above (Table 3). Other GOs (Supplementary Material 5) were less suggestive of likely organs, capabilities, or physiological processes of the syndermatan LCA. Nevertheless, neurogenesis, response to oxidative stress, regulation of acrosome reaction, etc. were almost certainly important for the syndermatan LCA, just as it is the case for so many other species.
The highest number of private transcript or protein clusters in A. vaga (Fig. 4A) as well as the highest total number of clusters in this species (Fig. 4B) add to previous evidence of genome duplication in the bdelloid stem line (Flot et al., 2013; Simion et al., 2021), which presumably was followed by sub- and neofunctionalization of part of the paralogues (Conant & Wolfe, 2008; Gleixner et al., 2012). The overall increased representation of A. vaga proteins in the 2114 core orthogroups compared to the other three syndermatan species further emphasizes this. However, the average numbers of transcripts per core orthogroup illustrate that the increase in A. vaga does not correspond to a doubling (Supplementary Material 5: 1.77 vs. 1.08–1.33), thus underscoring the degenerate state of tetraploidy in this bdelloid (Flot et al. 2013; Simion et al., 2021). In fact, gene loss might have occurred at elevated rates following genome duplication in the bdelloid stem line, due to relaxed functional constraint. Gene loss will also have played a major role in the evolution of Seisonidea and Acanthocephala (Mauer et al., 2021), as evidenced by the second smallest and smallest total numbers of protein clusters in S. nebaliae and P. laevis, respectively (Fig. 4B). Nevertheless, new genes could have originated on the lineage to the acanthocephalan to an increased extent, as already mentioned.
Conclusion
As shown above, the syndermatan LCA might have possessed a compact nuclear genome of about 50–100 Mb, of which about 40% or more was protein-coding. The presumably ~ 17,000 protein-coding genes should have had 4–6 exons and 3–5 introns on average. Considering respective data for other protostomes, a gene count as suggested here for the syndermatan LCA could reflect the loss of about 3000 genes since the split from the lineage to platyhelminths. Present results additionally indicate great divergence of monogonont, bdelloid, seisonid, and acanthocephalan genomes (Tables 1, 2). One explanation will be that presumably more than 500 million years elapsed since their LCA (Near et al., 1998; Herlyn, 2021). To put this into perspective, the ray-finned fish lineage (Actinopterygii) probably split from the lineage to lobe-finned vertebrates (Sarcopterygii) about 450 million years ago (Hedges, 2002). However, only about 50% or 69% of the genes of the pufferfish Takifugu rubripes (Temminck & Schlegel, 1850) have orthologous counterparts in extant sarcopterygian taxa, such as the chicken Gallus gallus (Linnaeus, 1758) (Hillier et al., 2004) and human (Homo sapiens Linnaeus, 1758) (Howe et al., 2016). In the syndermatans analyzed here, it has additionally to be considered that generation times are much shorter than in most vertebrates. Also, the reproductive strategies of monogononts, bdelloids, seisonids, and acanthocephalans greatly differ, thereby ranging from bisexuality in seisonids and acanthocephalans via metagenesis in monogononts to strict parthenogenesis in bdelloids (Wallace, 2002; Mark Welch & Meselson, 2003; Franch-Gras et al., 2019). Furthermore, syndermatan lifestyles cover free-living (Monogononta, Bdelloidea), epizoic/ectoparasitic way of living (Seisonidea), and endoparasitism with a complex lifecycle (Acanthocephala) (Herlyn, 2021). Yet, the loss of pathways and genes is a common pattern in the evolution of parasitism (e.g., Hahn et al., 2014; International Helminth Genomes Consortium, 2019; Mauer et al., 2021). Not least, genome duplication in the bdelloid stem line (Flot et al., 2013; Simion et al., 2021) could have fostered sub- and neofunctionalization due to relaxed functional constraint (Conant & Wolfe, 2008; Gleixner et al., 2012). Despite all these complicating factors, orthogroups containing transcripts or proteins of the four syndermatan species focused were found most numerous in clustering analysis (Fig. 4A). Upon expansion of the datasets there were still 194 clusters containing proteins conserved across the four syndermatan species analyzed, while lacking matches in both platyhelminths. The GOs represented in these clusters might point to the special relevance of the muscular pharynx or mastax and to the perception of light to the LCA of Syndermata, amongst others (Table 3; Supplementary Material 5).
Differences in gene and genome metrics as observed by us might reflect different qualities of the genome assemblies analyzed. For example, linkage of gene segments residing on different contigs may have gone undetected. This might especially have led to an underestimation of total CDS size and median CDS length in P. laevis, considering disproportionally long introns in the corresponding nuclear genome assembly (Table 2). On the other hand, metrics as observed here in P. laevis genes are not uncommon in parasites. For example, large introns such observed here in the acanthocephalan (median: 868 bp; mean: 1817 bp) have also been reported for the genome assemblies of two Digenea species (Platyhelminthes): the blood fluke Schistosoma mansoni Sambon, 1907 (mean intron length: 1692 bp) and the liver fluke Fasciola hepatica Linnaeus, 1758 (3700 bp), respectively (Cwiklinski et al., 2015; Chelomina, 2017). In addition, short CDSs and proteins as observed in P. laevis (Table 2) seem also to occur in Microsporidia (Katinka et al., 2001) and parasitic representatives of Tunicata (Berná & Alvarez-Valin, 2015). Correspondingly, we expect the combination of intron expansion and CDS shortening in P. laevis not to be an artifact.
Careful assembly and annotation of additional genomes of especially Acanthocephala and Seisonidea, but also Bdelloidea and Monogononta remain necessary to further shed light onto the evolution of genome size and structure within the Syndermata. The combination with additional methods for determining genome size, such as flow cytometry, could be advantageous here because repetitive regions are often underrepresented in genome assemblies, thereby shortening the assemblies compared to the genomes (see, e.g., Blommaert, 2020). Furthermore, additional complete genome assemblies will foreseeably become available with the increasing use of sequencing platforms (PacBio, Nanopore) enabling read lengths of several to many kilobases. Also, in-depth studies on seisonid lifestyles are desirable as food acquisition in Seisonidea species is still insufficiently elucidated to unambiguously classify them as parasites or commensals (compare Illgen, 1916; Segers & Melone, 1998; Ahlrichs & Riemann, 2018). With the present analysis, we believe to have come closer to the goal of understanding genome evolution within Syndermata.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. https://doi.org/10.1126/science.287.5461.2185.
Ahlrichs, W. H., 1997. Epidermal ultrastructure of Seison nebaliae and Seison annulatus, and a comparison of epidermal structures within the Gnathifera. Zoomorphology 117: 41–48. https://doi.org/10.1007/s004350050028.
Ahlrichs, W. H., & O. Riemann, 2018. Seisonidae—Miscellaneous invertebrates. De Gruyter: 55–86.
Barnett, D. W., E. K. Garrison, A. R. Quinlan, M. P. Strrömberg & G. T. Marth, 2011. Bamtools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27: 1691–1692. https://doi.org/10.1093/bioinformatics/btr174.
Berná, L. & F. Alvarez-Valin, 2015. Evolutionary volatile cysteines and protein disorder in the fast-evolving tunicate Oikopleura dioica. Marine Genomics 24: 47–54. https://doi.org/10.1016/j.margen.2015.07.007.
Blommaert, J., 2020. Genome size evolution: towards new model systems for old questions. Proceedings of the Royal Society B 287: 20201441. https://doi.org/10.1098/rspb.2020.1441.
Blommaert, J., S. Riss, B. Hecox-Lea, D. B. Mark Welch & C. P. Stelzer, 2019. Small, but surprisingly repetitive genomes: transposon expansion and not polyploidy has driven a doubling in genome size in a metazoan species complex. BMC Genomics 20: 466. https://doi.org/10.1186/s12864-019-5859-y.
Bolger, A. M., M. Lohse & B. Usadel, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. https://doi.org/10.1093/bioinformatics/btu170.
Brand, J. N., R. A. W. Wiberg, R. Pjeta, P. Bertemes, C. Beisel, P. Ladurner & L. Schärer, 2020. RNA-Seq of three free-living flatworm species suggests rapid evolution of reproduction-related genes. BMC Genomics 21: 462. https://doi.org/10.1186/s12864-020-06862-x.
Brown, P. & E. Walsh, 2019. Genome size and lifestyle in gnesiotrochan rotifers. Hydrobiologia 844: 105–115. https://doi.org/10.1007/s10750-018-3873-8.
Brůna, T., K. J. Hoff, A. Lomsadze, M. Stanke & M. Borodovsky, 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics and Bioinformatics 3: Iqaa108. https://doi.org/10.1093/nargab/lqaa108.
Buchfink, B., C. Xie & D. H. Huson, 2014. Fast and sensitive protein alignment using DIAMOND. Nature Methods 12: 59–60. https://doi.org/10.1038/nmeth.3176.
Byeon, E., B. S. Choi, J. C. Park, M. S. Kim, D. H. Kim, J. S. Lee, Y. H. Lee, et al., 2021. The genome of the freshwater monogonont rotifer Brachionus angularis: identification of phase I, II, and III detoxification genes and their roles in molecular ecotoxicology. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 38: 100821. https://doi.org/10.1016/j.cbd.2021.100821.
Chelomina, G. N., 2017. Genomics and transcriptomics of the Chinese liver fluke Clonorchis sinensis (Opisthorchiidae, Trematoda). Molecular Biology 51: 184–193. https://doi.org/10.1134/S0026893317020078.
Clément, P. & E. Wurdak, 1991. Rotifera. In Harrison, F. W. & E. E. Ruppert (eds), Microscopic Anatomy of Invertebrates, Aschelminthes, Vol. 4. Wiley-Liss Inc, New York: 219–297.
Conant, G. C. & K. H. Wolfe, 2008. Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics 9: 938–950. https://doi.org/10.1038/nrg2482.
Cwiklinski, K., J. P. Dalton, P. J. Dufresne, J. la Course, D. J. L. Williams, J. Hodgkinson & S. Paterson, 2015. The Fasciola hepatica genome: gene duplication and polymorphism reveals adaptation to the host environment and the capacity for rapid evolution. Genome Biology 16: 71. https://doi.org/10.1186/s13059-015-0632-2.
Dobin, A., C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson & T. R. Gingeras, 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. https://doi.org/10.1093/bioinformatics/bts635.
Dolezel, J., S. Bartos, H. Voglmayer & J. Greilhuber, 2003. Letter to the Editor-Nuclear DNA content and genome size of trout and human. Cytrometry Part A 51A: 127–128. https://doi.org/10.1002/cyto.a.10013.
Flot, J. F., B. Hespeels, X. Li, B. Noel, I. Arkhipova, E. G. J. Danchin, A. Hejnol, B. Henrissat, et al., 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500: 453–457. https://doi.org/10.1038/nature12326.
Fontaneto, D. & W. H. De Smet, 2015. Rotifera. In Schmidt-Rhaesa, A. (ed), Handbook of Zoology: Gastrotricha, Cycloneuralia and Gnathifera. De Gruyter, Berlin: 217–301.
Franch-Gras, L., C. Hahn, E. M. García-Roger, M. J. Carmona, M. Serra & A. Gómez, 2018. Genomic signatures of local adaptation to the degree of environmental predictability in rotifers. Scientific Reports 8: 16051.
Franch-Gras, L., E. Tarazona, E. M. García-Roger, M. J. Carmona, A. Gómez & M. Serrra, 2019. Rotifer adaptation to the unpredictability of the growing season. Hydrobiologia 844: 257–273. https://doi.org/10.1038/s41598-018-34188-y.
Francis, W. R. & G. Wörheide, 2017. Similar ratios of introns to intergenic sequence across animal genomes. Genome Biology and Evolution 9: 1582–1598. https://doi.org/10.1093/gbe/evx103.
Gabriel, L., K. J. Hoff, T. Brůna, M. Borodovsky, & M. Stanke, 2021. TSEBRA: transcript selector for BRAKER. bioRxiv preprint 2021.06.07. https://doi.org/10.1101/2021.06.07.447316.
Gilbert, W., 1978. Why genes in pieces? Nature 271: 501–501. https://doi.org/10.1038/271501a0.
Gleixner, E., H. Herlyn, S. Zimmerling, T. Burmester & T. Hankeln, 2012. Testes-specific hemoglobins in Drosophila evolved by a combination of sub- and neofunctionalization after gene duplication. BMC Evolutionary Biology 12: 34. https://doi.org/10.1186/1471-2148-12-34.
Goubert, C., L. Modolo, C. Vieira, C. ValienteMoro, P. Mavingui & M. Boulesteix, 2015. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Biology and Evolution 7: 1192–1205. https://doi.org/10.1093/gbe/evv050.
Grohme, M. A., S. Schloissnig, A. Rozanski, M. Pippel, G. R. Young, S. Winkler, H. Brandl, et al., 2018. The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature 554: 56–61. https://doi.org/10.1038/nature25473.
Hahn, C., B. Fromm & L. Bachmann, 2014. Comparative genomics of flatworms (Platyhelminthes) reveals shared genomic features of ecto- and endoparastic Neodermata. Genome Biology and Evolution 6: 1105–1117. https://doi.org/10.1093/gbe/evu078.
Hammond, R. A., 1966. The proboscis mechanism of Acanthocephalus ranae. Journal of Experimental Biology 45: 203–213. https://doi.org/10.1242/jeb.45.2.203.
Han, J., J. C. Park, B.-S. Choi, M.-S. Kim, H.-S. Kim, A. Hagiwara, H. G. Park, B.-Y. Lee & J.-S. Lee, 2019. The genome of the marine monogonont rotifer Brachionus plicatilis: genome-wide expression profiles of 28 cytochrome P450 genes in response to chlorpyrifos and 2-ethyl-phenanthrene. Aquatic Toxicology 214: 105230. https://doi.org/10.1016/j.aquatox.2019.105230.
Hecox-Lea, B. J. & D. B. Mark Welch, 2018. Evolutionary diversity and novelty of DNA repair genes in asexual bdelloid rotifers. BMC Evolutionary Biology 18: 177. https://doi.org/10.1186/s12862-018-1288-9.
Hedges, S. B., 2002. The origin and evolution of model organisms. Nature Reviews Genetics 3: 838–849. https://doi.org/10.1038/nrg929.
Herlyn, H., 2021. Thorny-headed worms (Acanthocephala): jaw-less members of jaw-bearing worms that parasitize jawed arthropods and jawed vertebrates. In de Baets, K. & J. W. Huntley (eds), The Evolution and Fossil Record of Parasitism. Topics in Geobiology, Vol. 49. Springer, New York: 273–313.
Herlyn, H. & U. Ehlers, 1997. Ultrastructure and function of the pharynx of Gnathostomula paradoxa (Gnathostomulida). Zoomorphology 117: 135–145. https://doi.org/10.1007/s004350050038.
Herlyn, H. & H. Taraschewski, 2017. Evolutionary anatomy of the muscular apparatus involved in the anchoring of Acanthocephala to the intestinal wall of their vertebrate hosts. Parasitology Research 116: 1207–1225. https://doi.org/10.1007/s00436-017-5398-x.
Herlyn, H., N. Martini & U. Ehlers, 2001. Organisation of the praesoma of Paratenuisentis ambiguus (Van Cleave, 1921) (Acanthocephala: Eoacanthocephala), with special reference to the lateral sense organs and musculature. Systematic Parasitology 50: 105–116. https://doi.org/10.1023/A:1011925516086.
Herlyn, H., O. Piskurek, J. U. Urgen Schmitz, U. Ehlers & H. Zischler, 2003. The syndermatan phylogeny and the evolution of acanthocephalan endoparasitism as inferred from 18S rDNA sequences. Molecular Phylogeny and Evolution 26: 155–164. https://doi.org/10.1016/S1055-7903(02)00309-3.
Hillier, L. W., W. Miller, E. Birney, W. Warren, R. C. Hardison, C. P. Ponting, P. Bork, et al., 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. https://doi.org/10.1038/nature03154.
Hoff, K. J., S. Lange, A. Lomsadze, M. Borodovsky & M. Stanke, 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32: 767–769. https://doi.org/10.1093/bioinformatics/btv661.
Hoff, K. J., A. Lomsadze, M. Borodovsky & M. Stanke, 2019. Whole-Genome Annotation with BRAKER. In Kollmar, M. (ed), Gene Prediction. Methods in Molecular Biology. Humana, New York: 65–95. https://doi.org/10.1007/978-1-4939-9173-0_5.
Howe, K., P. H. Schiffer, J. Zielinski, T. Wiehe, G. K. Laird, J. C. Marioni, O. Soylemez, F. Kondrashov & M. Leptin, 2016. Structure and evolutionary history of a large family of NLR proteins in the zebrafish. Open Biology Royal Society of London 6: 160009. https://doi.org/10.1098/rsob.160009.
Hur, J. H., K. van Doninck, M. L. Mandigo & M. Meselson, 2009. Degenerate tetraploidy was established before bdelloid rotifer families diverged. Molecular Biology and Evolution 26: 375–383. https://doi.org/10.1093/molbev/msn260.
Illgen, H., 1916. Zur Kenntnis der Biologie und Anatomie der parasitischen Rotatorienfamilie der Seisoniden. Zoologischer Anzeiger 47: 1–9.
International Helminth Genomes Consortium, 2019. Comparative genomics of the major parasitic worms. Nature Genetics 51: 163–174. https://doi.org/10.1038/s41588-018-0262-1.
Kang, H. M., M. S. Kim, B. S. Choi, D. H. Kim, H. J. Kim, U. K. Hwang, A. Hagiwara & J. S. Lee, 2020. The genome of the marine monogonont rotifer Brachionus rotundiformis and insight into species-specific detoxification components in Brachionus spp. Comparative biochemistry and physiology. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 36: 100714. https://doi.org/10.1016/j.cbd.2020.100714.
Katinka, M. D., S. Duprat, E. Cornillot, G. Metenier, F. Thomarat, G. Prensier, V. Barbe, E. Peyretaillade, P. Brottier, P. Wincker, F. Delbac, H. El Alaoui, P. Peyret, W. Saurin, M. Gouy, J. Weissenbach & C. P. Vivares, 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 412: 450–453. https://doi.org/10.1038/35106579.
Kaur, S., S. A. Stinson, G. C. diCenzo, 2022. Whole genome assemblies of Zophobas morio and Tenebrio molitor. bioRxiv. https://doi.org/10.1101/2022.12.21.521312
Kikuchi, T., J. A. Cotton, J. J. Dalzell, K. Hasegawa, N. Kanzaki, P. McVeigh, T. Takanashi, I. J. Tsai, S. A. Assefa, P. J. A. Cock, T. da Otto, M. Hunt, A. J. Reid, A. Sanchez-Flores, K. Tsuchihara, T. Yokoi, M. C. Larsson, J. Miwa, A. G. Maule, N. Sahashi, J. T. Jones & M. Berriman, 2011. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathogens 7(9): 1002219. https://doi.org/10.1371/journal.ppat.1002219.
Kim, H. S., J. Han, H. J. Kim, A. Hagiwara & J. S. Lee, 2017. Identification of 28 cytochrome P450 genes from the transcriptome of the marine rotifer Brachionus plicatilis and analysis of their expression. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 23: 1–7. https://doi.org/10.1016/j.cbd.2017.04.005.
Kim, H. S., B. Y. Lee, J. Han, C. B. Jeong, D. S. Hwang, M. C. Lee, H. M. Kang, D. H. Kim, H. J. Kim, S. Papakostas, S. A. J. Declerck, I. Y. Choi, A. Hagiwara, H. G. Park & J. S. Lee, 2018. The genome of the freshwater monogonont rotifer Brachionus calyciflorus. Molecular Ecology Resources 18: 646–655. https://doi.org/10.1111/1755-0998.12768.
Kim, D. H., M. S. Kim, A. Hagiwara & J. S. Lee, 2021. The genome of the minute marine rotifer Proales similis: genome-wide identification of 401 G protein-coupled receptor (GPCR) genes. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 39: 100861. https://doi.org/10.1016/j.cbd.2021.100861.
Leasi, F., G. W. Rouse & M. V. Sørensen, 2012. A new species of Paraseison (Rotifera: Seisonacea) from the coast of California, USA. Journal of the Marine 92: 959–965. https://doi.org/10.1017/S0025315411000129.
Li, H., B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis & R. Durbin, 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. https://doi.org/10.1093/bioinformatics/btp352.
Lomsadze, A., P. D. Burns & M. Borodovsky, 2014. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Research 42: e119. https://doi.org/10.1093/nar/gku557.
Mark Welch, D. B., 2000. Evidence from a protein-coding gene that acanthocephalans are rotifers. Invertebrate Biology 119: 17–26. https://doi.org/10.1111/j.1744-7410.2000.tb00170.x.
Mark Welch, D. B. & M. Meselson, 1998. Measurements of the genome size of the monogonont rotifer Brachionus plicatilis and of the bdelloid rotifers Philodina roseola and Habrotrocha constricta. Hydrobiologia 387(388): 395–402. https://doi.org/10.1023/A:1017063201343.
Mark Welch, D. B. & M. Meselson, 2003. Oocyte nuclear DNA content and GC proportion in rotifers of the anciently asexual Class Bdelloidea. Biological Journal of the Linnean Society 79: 85–91. https://doi.org/10.1046/j.1095-8312.2003.00180.x.
Mark Welch, J. L., D. B. Mark Welch & M. Meselson, 2004. Cytogenetic evidence for asexual evolution of bdelloid rotifers. Proceedings of the National Academy of USA 101: 1618–1621. https://doi.org/10.1073/pnas.0307677100.
Mark Welch, D. B., J. L. Mark Welch & M. Meselson, 2008. Evidence for degenerate tetraploidy in bdelloid rotifers. Proceedings of the National Academy of Sciences 105: 5145–5149. https://doi.org/10.1073/pnas.0800972105.
Mauer, K., S. L. Hellmann, M. Groth, A. C. Fröbius, H. Zischler, T. Hankeln & H. Herlyn, 2020. The genome, transcriptome, and proteome of the fish parasite Pomphorhynchus laevis (Acanthocephala). PLoS ONE 15: 1–30. https://doi.org/10.1371/journal.pone.0232973.
Mauer, K. M., H. Schmidt, M. Dittrich, A. C. Fröbius, S. L. Hellmann, H. Zischler, T. Hankeln & H. Herlyn, 2021. Genomics and transcriptomics of epizoic Seisonidea (Rotifera, syn. Syndermata) reveal strain formation and gradual gene loss with growing ties to the host. BMC Genomics 22: 1–24. https://doi.org/10.1186/s12864-021-07857-y.
Moran, J. V., R. J. DeBerardinis & H. H. Kazazian, 1999. Exon shuffling by L1 retrotransposition. Science 283: 1530–1534. https://doi.org/10.1126/science.283.5407.1530.
Near, T. J., J. R. Garey & S. A. Nadler, 1998. Phylogenetic relationships of the Acanthocephala inferred from 18S ribosomal DNA sequences. Molecular Phylogenetics and Evolution 10: 287–298. https://doi.org/10.1006/mpev.1998.0569.
Nowell, R. W., P. Almeida, C. G. Wilson, T. P. Smith, D. Fontaneto, A. Crisp, G. Micklem, A. Tunnacliffe, C. Boschetti & T. G. Barraclough, 2018. Comparative genomics of bdelloid rotifers: insights from desiccating and nondesiccating species. PLoS Biology Public Library of Science 16: e2004830. https://doi.org/10.1371/journal.pbio.2004830.
Nowell, R. W., C. G. Wilson, P. Almeida, P. H. Schiffer, D. Fontaneto, L. Becks, F. Rodriguez, I. R. Arkhipova & T. G. Barraclough, 2021. Evolutionary dynamics of transposable elements in bdelloid rotifers. eLife 10: 1–86. https://doi.org/10.7554/eLife.63194.
Ooms, J., 2022. rsvg: Render SVG Images into PDF, PNG, (Encapsulated) PostScript, or Bitmap Arrays.
Pagani, M., C. Ricci & C. A. Redi, 1993. Oogenesis in Macrotrachela quadricornifera (Rotifera, Bdelloidea)—I. Germarium eutely, karyotype and DNA content. Hydrobiologia 255: 225–230.
Parkinson, J., M. Mitreva, C. Whitton, M. Thomson, J. Daub, J. Martin, R. Schmid, et al., 2004. A transcriptomic analysis of the phylum Nematoda. Nature Genetics 36: 1259–1267. https://doi.org/10.1038/ng1472.
Patthy, L., 1999. Genome evolution and the evolution of exon-shuffling-a review. Gene 238: 103–114. https://doi.org/10.1016/S0378-1119(99)00228-0.
Pérez-Silva, J. G., M. Araujo-Voces & V. Quesada, 2018. Data and text mining nVenn: generalized, quasi-proportional Venn and Euler diagrams. Bioinformatics 34: 2322–2324. https://doi.org/10.1093/bioinformatics/bty109.
Perrot-Minnot, M. J., E. Guyonnet, L. Bollache & C. Lagrue, 2019. Differential patterns of definitive host use by two fish acanthocephalans occurring in sympatry: Pomphorhynchus laevis and Pomphorhynchus tereticollis. International Journal for Parasitology: Parasites and Wildlife 8: 135–144. https://doi.org/10.1016/j.ijppaw.2019.01.007.
Poulin, R. & H. S. Randhawa, 2015. Evolution of parasitism along convergent lines: From ecology to genomics. Parasitology 142(Suppl 1): S6–S15. https://doi.org/10.1017/S0031182013001674.
R Core Team, 2022. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
Rieger, R. & S. Tyler, 1995. Sister-group relationship of Gnathostomulida and Rotifera-Acanthocephala. Invertebrate Biology 114: 186–188. https://doi.org/10.2307/3226891.
Segers, H. & G. Melone, 1998. A comparative study of trophi morphology in Seisonidea (Rotifera). Journal of Zoology 244: 201–207. https://doi.org/10.1111/j.1469-7998.1998.tb00025.x.
Sielaff, M., H. Schmidt, T. H. Struck, D. Rosenkranz, D. B. Mark Welch, T. Hankeln & H. Herlyn, 2016. Phylogeny of Syndermata (syn. Rotifera): mitochondrial gene order verifies epizoic Seisonidea as sister to endoparasitic Acanthocephala within monophyletic Hemirotifera. Molecular Phylogenetics and Evolution 96: 79–92. https://doi.org/10.1016/j.ympev.2015.11.017.
Simion, P., J. Narayan, A. Houtain, A. Derzelle, L. Baudry, E. Nicolas, R. Arora, et al., 2021. Chromosome-level genome assembly reveals homologous chromosomes and recombination in asexual rotifer Adineta vaga. Science Advances 7: eabg4216. https://doi.org/10.1126/sciadv.abg4216.
Smit, A., & R. Hubley, 2008. RepeatModeler Open-1.0. 2008–2015.
Smit, A., R. Hubley, G. P. R., 2013. RepeatMasker Open-4.0. 2013–2015.
Sørensen, M. V. & G. Giribet, 2006. A modern approach to rotiferan phylogeny: combining morphological and molecular data. Molecular Phylogenetics and Evolution 40: 585–608. https://doi.org/10.1016/j.ympev.2006.04.001.
Sørensen, M. V., R. M. Kristensen & K. Worssae, 2016. The Gnathifera. Phyla Gnathostomulida, Rotifera (including Acanthocephala), and Micrognathozoa. In Brusca, R. C., W. Moore & S. M. Shuster (eds), Invertebrates. Sinauer Associates Inc, Sutherland: 613–434.
Spanu, P. D., J. C. Abbott, J. Amselem, T. A. Burgis, D. M. Soanes, K. Stüber, E. V. L. van Themaat, et al., 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546. https://doi.org/10.1126/science.1194573.
Stanke, M., O. Schöffmann, B. Morgenstern & S. Waack, 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7: 62. https://doi.org/10.1186/1471-2105-7-62.
Stanke, M., M. Diekhans, R. Baertsch & D. Haussler, 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. https://doi.org/10.1093/bioinformatics/btn013.
Stelzer, C. P., J. Blommaert, A. M. Waldvogel, M. Pichler, B. Hecox-Lea & D. B. Mark Welch, 2021a. Comparative analysis reveals within-population genome size variation in a rotifer is driven by large genomic elements with highly abundant satellite DNA repeat elements. BMC Biology 19: 1–17. https://doi.org/10.1186/s12915-021-01134-w.
Stelzer, C. P., M. Pichler & A. Hatheuer, 2021b. Linking genome size variation to population phenotypic variation within the rotifer, Brachionus asplanchnoidis. Communications Biology 4: 1–8. https://doi.org/10.1038/s42003-021-02131-z.
Struck, T. H., A. R. Wey-Fabrizius, A. Golombek, L. Hering, A. Weigert, C. Bleidorn, S. Klebow, N. Iakovenko, B. Hausdorf, M. Petersen, P. Kück, H. Herlyn & T. Hankeln, 2014. Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of Spiralia. Molecular Biology and Evolution 31: 1833–1849. https://doi.org/10.1093/molbev/msu143.
Taraschewski, H., 2015. Acanthocephala: functional morphology. In Schmidt-Rhaesa, A. (ed), Handbook of Zoology: Gastrotricha, Cycloneuralia and Gnathifera. De Gruyter, Berlin: 301–316.
Vakhrusheva, O. A., E. A. Mnatsakanova, Y. R. T. V. GalimovNeretina, E. S. Gerasimov, S. A. Naumenko, S. G. Ozerova, et al., 2020. Genomic signatures of recombination in a natural population of the bdelloid rotifer Adineta vaga. Nature Communications 11: 6421. https://doi.org/10.1038/s41467-020-19614-y.
von Haffner, K., 1950. Organisation und systematische Stellung der Acanthocephalen. Zool Anz. 145: 243–274.
Wallace, R. L., 2002. Rotifers: exquisite metazoans. Integrative and Comparative Biology 42: 660–667.
Wang, Y., D. Coleman-Derr, G. Chen & Y. Q. Gu, 2015. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research 43: 78–84. https://doi.org/10.1093/nar/gkv487.
Wang, S., S. Wang, Y. Luo, L. Xiao, X. Luo, S. Gao, Y. Dou, et al., 2016. Comparative genomics reveals adaptive evolution of Asian tapeworm in switching to a new intermediate host. Nature Communications 7: 12845. https://doi.org/10.1038/ncomms12845.
Wasik, K., J. Gurtowski, X. Zhou, O. M. Ramos, M. J. Delás, G. Battistoni, O. El Demerdash, et al., 2015. Genome and transcriptome of the regeneration-competent flatworm, Macrostomum lignano. Proceedings of the National Academy of Sciences USA 112: 12462–12467. https://doi.org/10.1073/pnas.1516718112.
Waterston, R. & J. Sulston, 1995. The genome of Caenorhabditis elegans. Proceedings of the National Academy of Sciences USA 92: 10836–10840. https://doi.org/10.1073/pnas.92.24.10836.
Wey-Fabrizius, A. R., H. Herlyn, B. Rieger, D. Rosenkranz, A. Witek, D. B. M. Welch, I. Ebersberger & T. Hankeln, 2014. Transcriptome data reveal syndermatan relationships and suggest the evolution of endoparasitism in Acanthocephala via an epizoic stage. PLoS ONE 9: e88618. https://doi.org/10.1371/journal.pone.0088618.
Wickham, H., 2016. ggplot2: Elegant Graphics for Data Analysis. Springer, New York.
Wickham, H., & D. Seidel, 2022. Scales: Scale Functions for Visualization.
Witek, A., H. Herlyn, A. Meyer, L. Boell, G. Bucher & T. Hankeln, 2008. EST based phylogenomics of Syndermata questions monophyly of Eurotatoria. BMC Evolutionary Biology 8: 345. https://doi.org/10.1186/1471-2148-8-345.
Xu, L., Z. Dong, L. Fang, Y. Luo, Z. Wei, H. Guo, G. Zhang, Y. Q. Gu, D. Coleman-Derr, Q. Xia & Y. Wang, 2019. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research 47: 52–58. https://doi.org/10.1093/nar/gkz333.
Yandell, M. & D. Ence, 2012. A beginner’s guide to eukaryotic genome annotation. Nature Reviews 13: 329–342. https://doi.org/10.1038/nrg3174.
Zrzavý, J., 2001. The interrelationships of metazoan parasites: a review of phylum-and higher-level hypotheses from recent morphological and molecular phylogenetic analyses. Folia Parasitologica 48: 81–103. https://doi.org/10.14411/fp.2001.013.
Acknowledgements
We gratefully acknowledge useful comments of two anonymous reviewers on a previous draft and financial support by Deutsche Forschungsgemeinschaft (HE3487/5-1).
Funding
Open Access funding enabled and organized by Projekt DEAL. Funding was provided by Deutsche Forschungsgemeinschaft (Grant no. HE3487/5-1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guest editors: Maria Špoljar, Diego Fontaneto, Elizabeth J. Walsh & Natalia Kuczyńska-Kippen / Diverse Rotifers in Diverse Ecosystems
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hagemann, L., Mauer, K.M., Hankeln, T. et al. Nuclear genome annotation of wheel animals and thorny-headed worms: inferences about the last common ancestor of Syndermata (Rotifera s.l.). Hydrobiologia 851, 2827–2844 (2024). https://doi.org/10.1007/s10750-023-05268-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05268-6