Abstract
Switzerland’s drainage systems are divided into three major European river basins, i.e. the Po, the Rhône, and the Rhine basins. Until recently 32 species of freshwater mussel species (i.e. belonging to the genera Anodonta and Unio) were recognized for the country, albeit their identity and number remain uncertain especially, given the recent recognition of Unio mancus Lamarck, 1819 from the Rhône in France, and Unio elongatulus C. Pfeiffer, 1825 and Anodonta exulcerata Porro, 1838 from the Po basin in Italy. In this study, we molecularly assess Swiss populations of freshwater mussels to understand the identity and number of species as well as to characterize their distributions within this geologically differentiated Alpine country. We collected 125 specimens in 42 lakes and rivers representing the three major basins and performed a phylogenetic investigation of the collected specimens using two mitochondrial markers (COI & 16S) and one nuclear marker (28S). COI Haplotype networks are then presented for the identified species. Our new findings show that Unio elongatulus inhabits water bodies north of the main Alpine arc. No living populations of Unio mancus could be detected in Switzerland. Anodonta exulcerata is recorded from two localities north of Lake Maggiore and in the Swiss part of Lake Lugano. Anodonta anatina (Linnaeus, 1758) shows genetic differences between southern alpine and northern alpine populations. Our genetic data from Swiss populations of unionid species provides new records and knowledge concerning freshwater mussels from Central Europe and specifically from the Alpine region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The hydrogeological network of Switzerland is connected to three major European river basins, i.e. the Po, the Rhône, and the Rhine basin. The latter two rivers have their sources and upper potamic habitats in the Swiss Alps. Both rivers play an important role in the water contribution to large parts of Europe. This makes the area a crucial ecological hub for freshwater flora and fauna, including 32 species of freshwater bivalves. Members of the family Unionidae Rafinesque, 1820 represent the largest freshwater mussels in body size. Unfortunately, public awareness is still practically nonexistent for the water cleansing benefits provided by these remarkable biofilterers.
Swiss naturalists were long fascinated by the autochthonous bivalve fauna; Studer (1789) recorded five species of unionid mussels for Switzerland, and later in 1820, he already listed six species of Unio, and two of Anodonta. In the following years, Swiss waterbodies were studied by numerous malacologists: Bourguignat (1862) added four Unio and eight Anodonta species from the Lake Lucerne describing several new species; Brot (1867) discussed the Unionidae living in Lake Geneva, and Godet (1911) reported on the forms of Unio crassus Philipsson, 1788 from Lake Neuchâtel. The investigations of all these authors were hindered by an increasingly chaotic system of names assigned to the level of species, subspecies, variety, forms etc., purely based on morphology of shells combined with hypothetical biogeographic consideration. The first to apply a more modern species concept was Schnitter (1922), who revised the unionid species of Switzerland and realised the importance of phenotypic plasticity (Ortmann, 1920), which had been the cause of an overestimation of the number of taxa. Despite this knowledge of the shell plasticity, the classical discrimination of taxa based on conchological characters, was continued for the next 40 years. Haas (1969) hypothesised the existence of hybrids in U. pictorum (Linnaeus, 1758) and U. mancus after investigating animals with ambiguous shell morphology. Later, enzyme variability analyses by Nagel and Badino (2001) lead to further clarification, rebutting Haas’ hypothesis. More advanced genetic methods (COI analysis) generated increasingly more molecular data whilst ushering Unionidae taxonomy into twenty first Century assessment standards (Lopes-Lima et al., 2017b; Pfeiffer et al., 2019). Currently, six species of Unionidae are recognized for Switzerland (Lopes‐Lima et al., 2017a). Since this last conservation assessment of European freshwater mussels, distinct Unio and Anodonta species have been recognized around Switzerland. Due to their ecological impact, it is important to monitor their presence in this small Alpine country.
This work’s primary aim was prompted by Froufe et al. (2017) upon the recovery of a cryptic species of Anodonta in Lake Lugano, northern Italy. This species was formally resurrected under the name Anodonta exulcerata Porro, 1838 by Riccardi et al. (2020). Since a section of Lake Lugano overlaps into Switzerland, detecting this species there became important for assessing other Swiss freshwater ecosystems. Unfortunately, the shell morphology of A. exulcerata does not allow a distinction from the similar A. anatina; however, A. exulcerata can be clearly identified using the conventional mitochondrial markers COI and 16S in a molecular phylogenetic analysis.
The second aim of this study is to investigate the Unio populations living in the catchment areas that flow into the Mediterranean, and if they have been able to cross the European watershed between the Rhône and the Rhine system? U. mancus and U. elongatulus were identified as target species. The first species was reported for northern Italy and the Ticino for many decades, however, Prié & Puillandre (2014) confirmed that U. elongatulus and U. mancus represent two separate species which can be distinguished using COI in a molecular analysis. Froufe et al. (2017) recognized that the historical records for”U. mancus “in northern Italy belong to U. elongatulus, which is a widespread unionid taxon in the area north of the Apennines to coastal Croatia (Lopes-Lima et al., 2017a).
The third aim of our investigation is to discover if living populations of U. mancus exist in the northwestern and central part of Switzerland. The species was reported from the Doubs in the French part of the Jura. The last records of living populations in the Swiss part of the Doubs were reported in 2003 (Rüetschi et al., 2010). The species’ known distribution covers Northern, Central and Southern France and Western and Southern Italy together with the Tyrrhenian Islands, Corsica, Sardinia and Sicily. However, a (re?)-dispersal along the Rhône system and potentially also the Doubs is theoretically given, particularly as the French freshwater canal system connects all larger rivers in eastern France. Specimens of Unio tumidus Philipsson, 1788 were included to represent the unionid fauna of the lakes in the Central Plateau of Switzerland.
To molecularly assess and delimit the selected Swiss species, we conducted phylogenetic analyses using two mitochondrial (COI, 16S) and one nuclear marker (28S) from 128 specimens culled from Swiss rivers and lakes. We enriched our dataset with sequences of the target species from areas outside Switzerland (Online Resource 4) to illustrate how Swiss species are nested in the larger context. Although a specimen of U. crassus was accidentally collected and added to the analysis to differentiate the trees. It was, however, not the target of this investigation.
Material and methods
Taxon sampling
Forty-two sites encompassing the major lakes and river systems of the Rhine, Rhône and Po basins were sampled during the years 2018–2020 (Fig. 1). One to eight individuals were collected at each site, resulting in a total of 125 specimens. To meet our research aim, only specimens from the target Unio species, i.e. U. mancus and U. elongatulus and U. tumidus were sampled. For Anodonta Lamarck, 1799, we included both species, Anodonta cygnea (Linnaeus, 1758) and A. anatina to detect possible A. exulcerata.
Concatenated phylogenetic tree obtained by Bayesian Inference (BI) analysis and Maximum Likelihood (ML) analysis of three genes (COI, 16S and 28S). ML was computed with RAxML and IQTree performing a standard bootstrap analysis. Support values indicated at the nodes. Subpopulations of A. anatina are labelled as A, B, C and D
The mussels were preserved on site in 100% ethanol. Preliminary determinations were made at the locality by the diver teams; specimens were then labelled and catalogued. All specimens are stored in 80% ethanol and preserved in the wet collection of the Natural History Museum Bern, Switzerland. In addition to the Swiss specimens, tissue snippets of 10 unionid specimens from the wet collection of the Senckenberg Museum Frankfurt (SMF), Germany, were added to the study. These snippets were supplied without taxonomic information, acting as a positive control for the extraction and PCR procedure performed in the laboratories at the NMBE (Online Resource 2).
A standardised field protocol was developed in cooperation with the SZKF (info fauna—Schweizerisches Zentrum für die Kartografie der Fauna (SZKF/CSCF)) by the research diver teams. Contained in the field log sheet is a complete set of physical, chemical and environmental data. These data include date, central coordinates, altitude, area searched, water temperature, general information on the dominant substrates and vegetation according to water depth, as well as information on the mussels observed, fish and other biota, with special attention to neozoa. This database will be available via the SZKF portal.
DNA extraction, markers description and amplification protocol
In a separate, sterile laboratory from that where the PCR is conducted, mantle snippets (somatic tissue) of approx. 5 mm3 size were cut with sterile surgical blades from each specimen. Somatic tissue in Unionidae contains (F) maternal mitochondria, and by clipping tissue from the mantle (F), mitochondria can be harvested (Froufe et al., 2016). Additional snippets were cut and stored for later research. Pre-extraction digestion was performed using the protocol of the Qiagen Blood and Tissue Kit (Qiagen cat. nr. 69506). Subsequent extraction was done by using a QIAcube extraction robot (DNeasy Blood Tissue and Rodent tails, standard protocol). Three markers commonly used in bivalves were selected: (1) the cytochrome c oxidase subunit I gene (COI; 710 bp amplicon length, LCO22/HCO700 primer pair) (Froufe et al., 2014); (2) the 16S ribosomal RNA gene (16S; 480 bp amplicon length, 16Sar/16Sbr primer pair) (Weigand et al., 2013) and (3) part of the 28S rRNA gene (28S; 810 bp amplicon length, RD1.3f/RD4b primer pair) (Whiting, 2002).
The PCR master mix consisted of 12.5 µl of GoTaq G2 HotStart Green Master Mix (Promega M7423), 6.5 µl nuclease free H2O (Sigma-Aldrich, W4502). For each reaction, 1 µl of each primer and 2 µl template DNA were added.
The PCR protocol for each marker described are as follows: For the LCO22/HCO700 (COI) primer pair the thermal regime begins with 3 min at 95 °C, followed by 35 cycles of 1 min at 95 °C, 1 min at 45 °C and 1 min at 72 °C and finishes at the end with 5 min at 72 °C. For the 16Sar/16Sbr (16S) primer pair, the conditions were 5 min at 95 °C in the first step, followed by 34 cycles of 30 s at 95 °C, 25 s at 52 °C and 45 s at 72 °C, and finally, 5 min at 72 °C. For the RD1.3f/RD4b (28S) primer pair, the protocol begins with 3 min at 95 °C, followed by 45 cycles of 30 s at 95 °C, 45 s at 50 °C and 60 s at 72 °C with a final elongation step of 5 min at 72 °C. PCR products were displayed together with a negative control and a 1000 bp ladder (BenchTop 100 bp DNA Ladder, G8291) in an agarose 1% gel for assessment of quality and primer efficiency. LGC (LGC Genomics Berlin) performed PCR product purification and sequencing with the above primers. The generated sequence data are downloadable from GenBank (Online Resource 2).
Phylogenetic analyses and species delimitation
From the initial 135 specimens, 128 were successfully sequenced for the analyses. The AB1 sequences were processed in Geneious Ver.9.1.8 (Biomatters Ltd.). Every sequence was verified through a BLAST analysis (National Center for Biotechnology Information (NCBI)). Wherever possible, we focused on BLAST results, providing vouchers from the latest publications involving Unionidae e.g., A. exulcerata MF414281 (Froufe et al. 2017). For comparing the Swiss data in a broader geographical context, a selection of sequences from the latest publications (Froufe et al., 2017; Klishko et al., 2018; Marrone et al. 2019; Lopes-Lima et al., 2021) were added to the phylogenetic analysis (Online Resource 2). The final sample size for a three-marker approach (COI, 16S, 28S) is 172 and the sample size for the ASAP analysis consisted of 246 COI sequences.
For the alignment, the MAFFT v.7.222 plugin of Geneious (Katoh & Standley, 2013) was implemented. The L- INS-i algorithm and the 1PAM/k = 2 matrix chosen for the alignment construction. Every gene alignment was verified and, when needed, improved by excluding positions of unreliable parts of the alignment. The final alignment length after editing was as follows (original first MAFFT alignment length in brackets): COI 654 bp (702 bp), 16S 497 bp (539 bp) and 28S 644 bp (827 bp) summing up to a total of 1795 bp in the concatenated alignment block. The final alignment length for the COI phylogeny was 546 bp. Substitution saturation was assessed using Xia’s test in DAMBE v.7.058 with default settings (Xia et al., 2003; Xia, 2017, 2018).
Species partitions analysis was performed on the Assemble Species by Automatic Partitioning (ASAP) web tool (https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html) (Puillandre et al., 2021), using the Kimura (K80) (Kimura, 1980) substitution model with the settings for the transition/transversion rate ratio (ts/tv) = 2 and the standard advanced options for 5 best partition suggestions for 242 COI sequences, excluding Potomida littoralis (Cuvier, 1798), Microcondylaea bonellii (Férussac, 1827) and Sinanodonta woodiana (I. Lea, 1834) from the analysis.
The protein coding COI fragment was defined in two partitions: the first two codon positions as one partition and the third codon position as a second. Both 16S and 28S alignment data sets were not partitioned and were handled each as a single group for the analyses. The final data block contained 172 individuals. Partitionfinder Ver. 2.1 (Lanfear et al., 2012, 2017) was implemented using the greedy algorithm settings to look for the optimal evolutionary models for the partitions and analysis methods through the corrected Akaike Information Criterion (AICc).
Maximum Likelihood (ML) analysis was computed with RAxML (Stamatakis, 2014) and IQTree (Nguyen et al., 2015; Chernomor et al., 2016; Minh et al., 2020) setting GTR + I + G as substitution model. For RAxML, the settings were set for a rapid bootstrap analysis with 2000 replicates in a single run. For the IQTree analysis a standard bootstrap analysis was performed with the settings of 100 bootstrap alignments and 1000 replicates for the SH-aLRT (approximate likelihood ratio test (aLRT) and Shimodaira–Hasegawa (SH)) (Guindon et al., 2010; Anisimova et al., 2011).
Bayesian Inference (BI) was performed using MrBayes v3.2.2 × 64 (Huelsenbeck & Ronquist, 2001; Ronquist & Huelsenbeck, 2003; Ronquist et al., 2012). The substitution model was chosen according to the Partitionfinder definition: the best model for COI and 16S was GTR + I + G and for 28S GTR + G. The settings for the Monte Carlo Markov Chain (MCMC) parameter were as follows: four chains and four separate runs for 20 × 106 generations, temperature set to 0.1, with a tree sampling frequency of 1000 and a burn in of 25%. For convergence diagnostics, Tracer (Rambaut et al., 2018) was implemented to analyse the effective sample size (ESS).
Tree display and editing was performed using FigTree v1.4.4 (https://github.com/rambaut/figtree/releases). MrBayes and RAxML were performed through UBELIX (http://www.id.unibe.ch/hpc), the HPC cluster at the University of Bern. The IQTree analysis was performed through the IQ-Tree web server http://iqtree.cibiv.univie.ac.at/. The haplotype networks were built with Popart v.1.7 (Leigh & Bryant, 2015) (http://popart.otago.ac.nz), using the TCS algorithm (Clement et al., 2002).
Results
Sequence analyses and species delimitation
Sequence numbers obtained per species are given in Online Resource 1. The substitution saturation assessment for the sequences using Xia’s test showed no signs of saturation (P = 0.00). Tracer analysis of the BI run showed no anomaly in the ESS scores (> 200). ASAP suggested one result out of 5 possible partition solutions, it consists of 9 groups in 242 specimens (Online Resource 3).
Phylogeny and distribution patterns in Switzerland
The concatenated phylogenetic tree shown in Fig. 2 is a combination of the Bayesian Inference (BI) and the Maximum Likelihood analysis (RAxML & IQTree) for a dataset of 172 specimens. It shows high support for the clades in the tree topology in that Unio and Anodonta are monophyletic. The U. tumidus clade splits early from the greater Unio cluster as shown in Fig. 2. The U. crassus group splits before the U. pictorum/U. mancus/U. elongatulus cluster (Fig. 3). The Anodonta clade splits in the three Anodonta groups (Fig. 2): A. exulcerata is sister group to A. cygnea, the A. anatina cluster is the counterpart to this group. The topology of the A. anatina cluster suggests four branches, with only one being supported by the node values: group A, consisting of Iranoturanic individuals (Fig. 4).
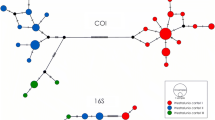
The haplotype network for U. elongatulus (Fig. 5) shows that the Swiss Rhine population shares the same haplotype as the populations from the Padano-Adriatic basins (Po, Reno) (Marrone et al., 2019). In Fig. 6, the haplotype network for A. anatina shows three distinguishable haplogroups displaying the topology shown in the phylogenetic tree. In Fig. 7, Anodonta cygnea displays one central haplogroup (central European), one Italian/Iberian and two distinct Turkish haplotypes. In Fig. 8, A. exulcerata shows 2 Croatian haplotypes, a central Italian/Swiss haplogroup and an Italian haplotype. The haplotypes with the corresponding sequences are given in Online Resource 1.
Discussion
BLAST and ASAP species delimitation
As stated by Froufe et al. (2017), an identification of species using morphological traits is rather difficult in the genus Unio. This pattern is in line with the display of high plasticity witnessed in shell morphology in members of Anodontini Rafinesque, 1820 (Zieritz & Aldridge, 2009; Zieritz et al., 2010; Riccardi et al., 2020). This situation is also known for other freshwater bivalves as well, e.g. Hyriidae Swainson, 1840 (Baker et al., 2003; Yusseppone et al., 2018). The BLAST results and ASAP partitioning facilitates the assignment of the individuals to their respective species identification.
Our investigation could not reveal any living population of U. mancus in Switzerland. However, U. elongatulus specimens from Switzerland could be confirmed by BLAST. Six specimens were first identified as U. tumidus in the field.
Anodonta exulcerata specimens however, do occur in the Swiss sampling shown in BLAST. The Swiss population belongs to the Italian haplogroup AE03 (Froufe et al., 2017) (Ticino/Lake Maggiore/Po) as shown in Fig. 8. Despite the variance displayed in the haplotype network, the ASAP analysis classified all the A. anatina individuals to one group. The Ticino population (AA01 and AA02) is geographically and genetically distinct from all other Swiss A. anatina populations (Fig. 6). In contrast, the Rhine basin populations form a fragmented pattern throughout the Swiss water systems. The Rhine population shares on the one side, haplotypes with eastern Russian and Ukrainian populations (AA12 and AA144). The west European (AA03, AA04 and AA06) haplogroups include Portuguese, Spanish, French, Czech, Italian and West-Ukrainian specimens. This pattern could be explained by multiple refugia for A. anatina during the last glacial periods (Froufe et al., 2014).
The topology in the haplotype network of A. cygnea shows a central homogenous haplogroup (AC02). Although the additional sequences used for the haplotype network cover Croatia, Sweden, Poland and France in the haplogroup (AC02), no higher diversity is indicated as observed in A. anatina.
Molecular phylogeny
In the concatenated phylogeny (COI, 16S and 28S) in Fig. 2, the node splitting Unio and Anodonta is supported with a posterior probability of BI = 1 and bootstrap values of BS = 99% and BS = 99%, with P. littoralis and M. bonelli changing position close to the Unio clade. The Position of Gonideinae Ortmann, 1916 is different in Froufe (2017: 3266, Fig. 6), which is explainable with the greater data set used in the analysis. For the species clades, the topology is coincident with that of Lopes-Lima et al. (2017b), Froufe et al. (2017), Marrone et al. (2019) and Riccardi et al. (2020).
Anodonta exulcerata
The haplotype network shows that AnoExu1 (Froufe et al., 2017) forms a haplogroup with the Swiss group, defining it as the Lake Maggiore haplotype. There were no geographical indications on AnuExu1 in Froufe et al. (2017). We identified one additional A. exulcerata, NMBE 565817 (Ceresio, Agno Camping) in the concatenated phylogeny. However, due to the missing COI sequence, it was not possible to detect it in the preceding BLAST analysis. The three A. exulcerata individuals from Switzerland originate from a rather small pond, Laghetto Demanio in Gudo, which is situated north of Lake Maggiore, upstream of the Ticino River in the Magadino plain. The pond constitutes a remnant tributary before the river was canalized. Thus, the enclosed fauna represents a population isolated from the canalised mainstream. Anodonta exulcerata NMBE 565817 originates from Lake Lugano, which is connected via the River Tresa to Lake Maggiore (Fig. 9). Riccardi’s (2020: 3, Table 1) sampling at Lake Lugano resulted in four A. cygnea specimens. These findings imply that A. exulcerata has a wider distribution in the southern Swiss and northern Italian lakes and rivers. Further investigation of the surrounding ponds and tributary remnants could well harbour additional populations warranting future consideration.
Unio mancus
The species has been split into three subspecies: U. mancus mancus, U. mancus requienii Michaud, 1831 and U. mancus turtonii Payraudeau, 1826. Froufe et al. (2017) found that the latter two subspecies only live in affluents of western Italy and considered uniting them into a single subspecies (i.e., U. mancus turtonii). In this case, Unio mancus mancus is restricted to the western region of France, Pyrenees and eastern Spain. This hypothesis was recently refuted by Marrone et al. (2019) due to anthropogenic influence in the Tyrrhenian drainage basin such that vicariant speciation of U. mancus requienii could not be tested. Moreover, the COI haplotype network in Marrone et al. (2019: 347, Fig. 7) used a broader COI dataset, showing well-defined haplotypes for all three subspecies. In addition, Prié & Puillandre (2014) show well-supported subspecies clades in Fig. 2 (COI, 16S, 28S) and in the Online Resource 4 (16S). Despite the same genetic markers, the presented concatenated phylogenetic tree did not show any U. mancus from Switzerland. When we revisited their sampling localities at the Swiss Doubs, only old shell fragments and no living Unio sp. individuals were found. However, the voucher collection in NMBE houses fresh shells collected in the mid 1980’s from the Doubs near St. Ursanne, indicating that the species has recently been exterminated. Additional sampling should focus on Lake Geneva, which potentially harbours this species as well.
Unio elongatulus
The distribution of U. elongatulus spans from north of the Apennines to the coastal region of Croatia (Froufe et al., 2017). The new records of U. elongatulus north of the Alps extend the northern distribution range of this species (Fig. 10). Moreover, since the populations occur scattered all over central and north-eastern Switzerland, no biogeographical pattern can be detected. This probably results from stocking events of potential host fish species throughout Switzerland. Such human mediated transport can easily move glochidia via infected host fishes northwards over the central Alpine chain. Host fishes for U. elongatulus include salmonids, freshwater blennies and cyprinids (Prié et al., 2012). Anthropogenic interference involving similarly infected host fishes with U. elongatulus and S. woodiana has also been reported in Florence, Italy (Froufe et al. 2017).
Our U. elongatulus samples cluster together with the neotype MG967432 from Vipava (= Vipacco) River near Gabria Inferiore (Marrone et al., 2019), thus confirming the correct species identification. The U. elongatulus specimens do not show any subclade formations in the single gene and three-marker phylogenetic tree. The haplotype network shows that the Rhine population is part of a haplotype with the Padano—Adriatic basins’ specimens (Marrone et al., 2019). Consequently, the entire U. elongatulus sampling from Switzerland originates from the southern alpine region. This opens new dispersal possibilities for this species towards Germany via the Rhine River such that the entire Swiss Rhine drainage system can be expected to be populated by U. elongatulus in near future.
The non-target taxa for this investigation showed interesting topologies in the phylogenetic analyses. Although not part of the main investigation, they are briefly pointed out here.
Anodonta cygnea
The low genetic variation A. cygnea observed in the single gene phylogeny and haplotype network does not change with additional molecular data. The clade is still compact and short-branched, which is a contrast to the higher variation shown in A. anatina. Possible explanations for this low variability could be that either the distribution of the individuals or certain populations appeared recently or there is anthropogenic distribution involving infected host fish distribution. The haplotype network shows no variation. This may also be an artefact of the sequence choice and certainly requires a larger data set with a broader geographical scope.
Anodonta anatina
The A. anatina clade indicates a topology with a partitioning into three subclades, but without full support in the node values. The subpopulations in Fig. 4 are congruent with the haplogroups depicted in Fig. 6. The results of the phylogenetic analysis present a similar topology as shown by Froufe et al. (2014: 7, Fig. 2), where the main cluster is partitioned into a clade encompassing the Ebro and Adriatic basins, a pan-European clade and the Iberian clades. The partitioning into these clades was to be expected ie. recently, a Transbaikal distribution of A. anatina was reported showing the inclusion of Ukrainian and East-Russian individuals into the pan-European A. anatina haplogroups (Klishko et al., 2018). However, partition A, the Turanic partition, is distinctly separated from the main A. anatina cluster with high node support in the phylogenetic tree. In the haplotype network, the AA17 haplogroup separates from that of the Ukrainian (AA16) by 10 mutations.
Partition B individuals of A. anatina (Lake Maggiore) were labelled during the field work as Anodonta sp.. BLAST analysis identified them as closely related to the A. anatina specimens, which belong to the biogeographical clade of the Italian/Ebro group (Froufe et al., 2014). In the haplotype network, they form the haplogroup consisting of AA01 and AA02 and are separated by 12 mutations.
Partition C in the A. anatina clade consists of the haplotypes AA11 to AA16. The East-Russian individuals originate from the Lake Baikal area. Together with the oriental individuals (AA10), it may be considered the EU-Oriental-Russian group. These specimens were part of the broader EU haplotype group in Klishko (2018: 7, Fig. 6).
Partition D is represented by the haplotypes AA03 to AA09. Individuals originate from West-Ukraine (AA07) over the Rhine basin into France (AA04), forming the Pan-European group.
By implementing a larger dataset in future investigations, the relationships between the different European groups in the A. anatina clade can likely be resolved.
Unio tumidus
Unio tumidus was chosen to support the structuring of the phylogenetic and ASAP analyses. In the phylogenetic tree it forms a well-defined compact clade, separating U. tumidus from U. crassus and U. elongatulus with high support (BI = 1; BS = 97%; BS = 94%). Future investigations covering a pan European context will do well to integrate Swiss U. tumidus genetic data derived from this work.
Conclusions
In our study, we present significant genetic data that enhances the known distribution of the family Unionidae in Central European freshwater systems. Of the 120 Swiss individuals, 46 specimens belong to A. anatina, 21 to A. cygnea, 4 to A. exulcerata, 1 to U. crassus, 19 to U. tumidus and 29 U. elongatulus. This information is listed together with the coordinates and GenBank accession numbers in the Online Resource 2.
We show that Anodonta exulcerata occurs in southern Switzerland along the River Ticino and is likely to inhabit other lakes and rivers in the area. To adequately assess the distribution of A. exulcerata, more lakes in the Tessin region need to be investigated. Although Unio mancus was not verified in Switzerland, further investigations are needed in the Swiss Rhône, the Lake Geneva and the Doubs system to detect cryptic populations. We recorded U. elongatulus from several sites north of the Alps, which proved to be closely related to those found in northern Italy. This is an alarming finding that has up to now not been reported. As this new invasion of U. elongatulus jeopardizes the ecology of the Rhine drainage system, the biome inhabited by indigenous species will drastically change, shifting the IUCN threatened status more towards extinction. In addition, our findings indicate that the A. anatina records in Switzerland show a north to south and eastern distribution pattern, and that Anodonta cygnea is the second most identified Anodonta species in the sampling.
The ecological importance of freshwater bivalves for river and lake systems is evident: being filter feeders, freshwater mussels greatly improve the water quality. A healthy mussel population in a river can retain up to 50% of the seston (Pusch et al., 2001). The size of the filtered particles differs for example between the species: A. anatina and U. pictorum retain particle sizes from 4 µm upwards and certain North American species filter particles smaller than 1 µm (Prié, 2017). Therefore, high biodiversity in freshwater mussel populations is a priority. The Red List for the molluscs of Switzerland (Rüetschi et al., 2010) listed a concerning number of large freshwater mussel species as threatened: Anodonta anatina VU; Unio crassus; Unio mancus EN; Unio pictorum EN; Unio tumidus. Specifically, Unio crassus in Switzerland has shown how a once widespread species rapidly suffered population decline since the 1950s. The species became almost extinct due to intensive farming and the introduced muskrat (Ondatra zibethicus (Linnaeus, 1766)) (Vicentini & Pfändler, 2001; Vicentini, 2004; Schwarzer & Neubert, 2014; Carlevaro & Stucki, 2020). Today, there are less than ten known live populations of U. crassus in Switzerland (Rüetschi et al., 2010). Current measures to protect the highly vulnerable populations of U. crassus showed promising results in the Canton of Zürich. One declining population could be stabilized with the method of ex situ infection of host fishes (Vicentini, 2006; Schwarzer, 2021). Despite all efforts, one of the three last known populations in the Canton of Zürich got lost (Isabelle Flöss, personal communication, July 21, 2021). Our scientific investigation widens the knowledge of large, Swiss indigenous freshwater bivalves. The genetic data is paramount for continuing the protection and monitoring of these important mussel species. In light of agricultural pollution, these natural water filters deliver an exceptional ecosystem service. Lastly, the data collected during this study will serve well to improve the classification of unionid taxa recorded in the IUCN Red List for Switzerland (Rüetschi et al., 2010).
Data availability
Sequence data are downloadable from GenBank (https://www.ncbi.nlm.nih.gov/genbank/), species distribution data are downloadable from CSCF database (http://www.cscf.ch/cscf/de/home.html)
Code availability
Not applicable.
References
Anisimova, M., M. Gil, J.-F. Dufayard, C. Dessimoz & O. Gascuel, 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Systematic Biology 60: 685–699.
Baker, A. M., C. Bartlett, S. E. Bunn, K. Goudkamp, F. Sheldon & J. M. Hughes, 2003. Cryptic species and morphological plasticity in long-lived bivalves (Unionoida: Hyriidae) from inland Australia. Molecular Ecology 12: 2707–2717.
Brot, A., 1867. Étude sur les coquilles de la famille des Nayades qui habitent le bassin du Léman. Association zoologique de Léman année 1866, 55 Seiten
Bourguignat, J. R., 1862. Malacologie du lac des Quatre-Cantons et de ses environs. Revue et Magasin de Zoologie, (2) 14 [1862] (11): 430–444, (12): 465–477, pl. 18–21 [pl. 18 –19 datées 1863], 15 [1863] (1): 5–26. Paris. [= pp. I–VIII, 9–72, pl. 1–4, Paris (J.–B. BAILLIÈRE), publié tout entier en Novembre 1862.]
Carlevaro, A., & P. Stucki, 2020. Kurzbericht: genetische Charakterisierung von dreizehn Bachmuschel-Populationen (Unio crassus) in der Schweiz
Chernomor, O., A. von Haeseler & B. Q. Minh, 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008.
Clement, M., Q. Snell, P. Walke, D. Posada, & K. Crandall, 2002. TCS: estimating gene genealogies. In Proceedings 16th International Parallel and Distributed Processing Symposium. IEEE, Ft. Lauderdale, FL: 7 pp, http://ieeexplore.ieee.org/document/1016585/.
Froufe, E., H. M. Gan, Y. P. Lee, J. Carneiro, S. Varandas, A. Teixeira, A. Zieritz, R. Sousa & M. Lopes-Lima, 2016. The male and female complete mitochondrial genome sequences of the Endangered freshwater mussel Potomida littoralis (Cuvier, 1798) (Bivalvia: Unionidae). Mitochondrial DNA Part A 27: 3571–3572.
Froufe, E., M. Lopes-Lima, N. Riccardi, S. Zaccara, I. Vanetti, J. Lajtner, A. Teixeira, S. Varandas, V. Prié, A. Zieritz, R. Sousa & A. E. Bogan, 2017. Lifting the curtain on the freshwater mussel diversity of the Italian Peninsula and Croatian Adriatic coast. Biodiversity and Conservation 26: 3255–3274.
Froufe, E., C. Sobral, A. Teixeira, R. Sousa, S. Varandas, D. C. Aldridge & M. Lopes-Lima, 2014. Genetic diversity of the pan-European freshwater mussel Anodonta anatina (Bivalvia: Unionoida) based on CO1: new phylogenetic insights and implications for conservation: GENETIC DIVERSITY ANODONTA ANATINA EUROPE. Aquatic Conservation 24: 561–574.
Godet, P., 1911. Contribution à l’ histoire naturelle des Najades Suisses. Unio consetaneus et ses variétés neuchâteloises. Bulletin de la Société Neuchâteloise des Sciences Naturelles, 38, 178
Guindon, S., J.-F. Dufayard, V. Lefort, M. Anisimova, W. Hordijk & O. Gascuel, 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321.
Huelsenbeck, J. P. & F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755.
Katoh, K. & D. M. Standley, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780.
Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120.
Klishko, O. K., M. Lopes-Lima, A. E. Bogan, D. V. Matafonov & E. Froufe, 2018. Morphological and molecular analyses of Anodontinae species (Bivalvia, Unionidae) of Lake Baikal and Transbaikalia. PLOS ONE 13: e0194944.
Lanfear, R., B. Calcott, S. Y. W. Ho & S. Guindon, 2012. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701.
Lanfear, R., P. B. Frandsen, A. M. Wright, T. Senfeld & B. Calcott, 2017. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773.
Leigh, J. W. & D. Bryant, 2015. Popart: full-feature software for haplotype network construction. Methods in Ecology and Evolution Wiley Online Library 6: 1110–1116.
Lopes-Lima, M., R. Sousa, J. Geist, D. C. Aldridge, R. Araujo, J. Bergengren, Y. Bespalaya, E. Bódis, L. Burlakova, D. Van Damme, K. Douda, E. Froufe, D. Georgiev, C. Gumpinger, A. Karatayev, Ü. Kebapçi, I. Killeen, J. Lajtner, B. M. Larsen, R. Lauceri, A. Legakis, S. Lois, S. Lundberg, E. Moorkens, G. Motte, K.-O. Nagel, P. Ondina, A. Outeiro, M. Paunovic, V. Prié, T. von Proschwitz, N. Riccardi, M. Rudzīte, M. Rudzītis, C. Scheder, M. Seddon, H. Şereflişan, V. Simić, S. Sokolova, K. Stoeckl, J. Taskinen, A. Teixeira, F. Thielen, T. Trichkova, S. Varandas, H. Vicentini, K. Zajac, T. Zajac & S. Zogaris, 2017a. Conservation status of freshwater mussels in Europe: state of the art and future challenges. Biological Reviews 92: 572–607.
Lopes-Lima, M., E. Froufe, Tu. V. Do, M. Ghamizi, K. E. Mock, Ü. Kebapçi, O. Klishko, S. Kovitvadhi, U. Kovitvadhi, O. S. Paulo, J. M. Pfeiffer III., M. Raley, N. Riccardi, H. Şereflişan, R. Sousa, A. Teixeira, S. Varandas, X. Wu, D. T. Zanatta, A. Zieritz & A. E. Bogan, 2017b. Phylogeny of most species rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): defining modern subfamilies and tribes. Molecular Phylogenetics and Evolution 106: 174–191.
Lopes-Lima, M., M. E. Gürlek, Ü. Kebapçı, H. Şereflişan, T. Yanık, A. Mirzajani, E. Neubert, V. Prié, A. Teixeira, A. Gomes-dos-Santos, D. Barros-García, I. N. Bolotov, A. V. Kondakov, I. V. Vikhrev, A. A. Tomilova, T. Özcan, A. Altun, D. V. Gonçalves, A. E. Bogan & E. Froufe, 2021. Diversity, biogeography, evolutionary relationships, and conservation of Eastern Mediterranean freshwater mussels (Bivalvia: Unionidae). Molecular Phylogenetics and Evolution 163: 107261.
Marrone, F., G. Nardi, S. Cianfanelli, M. Govedič, S. A. Barra, M. Arculeo & M. Bodon, 2019. Diversity and taxonomy of the genus Unio Philipsson in Italy, with the designation of a neotype for Unio elongatulus C. Pfeiffer (Mollusca, Bivalvia, Unionidae). Zootaxa 4545: 339.
Minh, B. Q., H. A. Schmidt, O. Chernomor, D. Schrempf, M. D. Woodhams, A. von Haeseler & R. Lanfear, 2020. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution 37: 1530–1534.
Nagel, K.-O., & G. Badino, 2001. Population Genetics and Systematics of European Unionoidea. Ecology and Evolution of the Freshwater Mussels Unionoida. Springer, Berlin pp. 51–80.
Nguyen, L.-T., H. A. Schmidt, A. von Haeseler & B. Q. Minh, 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274.
Ortmann, A. E., 1920. Correlation of shape and station in fresh-water mussels (Naiades). Proceedings of the American Philosophical Society 59: 269–312.
Prié, V., 2017. Naïades et autres bivalves d’eau douce de France. Biotope éditions.
Prié, V. & N. Puillandre, 2014. Molecular phylogeny, taxonomy, and distribution of French Unio species (Bivalvia, Unionidae). Hydrobiologia.
Prié, V., N. Puillandre, & P. Bouchet, 2012. Bad taxonomy can kill: molecular reevaluation of Unio mancus Lamarck, 1819 (Bivalvia : Unionidae) and its accepted subspecies. 18.
Puillandre, N., S. Brouillet & G. Achaz, 2021. ASAP: assemble species by automatic partitioning. Molecular Ecology Resources 21: 609–620.
Pusch, M., J. Siefert & N. Walz, 2001. Filtration and respiration rates of two Unionid species and their impact on the water quality of a lowland river. In Bauer, G. & K. Wächtler (eds), Ecology and Evolution of the Freshwater Mussels Unionoida. Springer, Berlin: 317–326.
Rambaut, A., A. J. Drummond, D. Xie, G. Baele & M. A. Suchard, 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology 67: 901–904.
Riccardi, N., E. Froufe, A. E. Bogan, A. Zieritz, A. Teixeira, I. Vanetti, S. Varandas, S. Zaccara, K.-O. Nagel & M. Lopes-Lima, 2020. Phylogeny of European Anodontini (Bivalvia: Unionidae) with a redescription of Anodonta exulcerata. Zoological Journal of the Linnean Society 189: 745–761.
Ronquist, F. & J. P. Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Ronquist, F., M. Teslenko, P. van der Mark, D. L. Ayres, A. Darling, S. Höhna, B. Larget, L. Liu, M. A. Suchard & J. P. Huelsenbeck, 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
Rüetschi, J., P. Stucki, P. Müller, H. Vicentini, & F. Claude, 2010. Rote Liste Weichtiere (Schnecken und Muscheln). Gefährdete Arten der Schweiz, Stand 2010.
Schwarzer, A. 2021. Wiederansiedlung der Bachmuschel (Unio crassus) im Kanton Aargau – ein Projekt der Sektion Jagd und Fischerei. https://www.aschwarzer.net/bachmuschel-2.
Schwarzer, A., & E. Neubert, 2014. Die Grossmuscheln im Kanton Solothurn, unter besonderer Berücksichtigung der Bachmuschel Unio crassus Philipsson 1788. Naturforschende Gesellschaft des Kantons Solothurn, Mitteilungen 10.
Schnitter, H., 1922. Die Najaden der Schweiz mit besonderer Berücksichtigung der Umgebung von Basel. Dissertation Universität Basel, 200 Seiten + Anhang
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313.
Vicentini, H., 2004. Bachmuscheln (Unio crassus) in Seen der Schweiz. Lauterbornia 50: 31–38.
Vicentini, H., 2006. Ansiedlungsversuche Bachmuschel (Unio crassus) im Kanton Aargau, 2006.
Vicentini, H. & U. Pfändler, 2001. Die Bachmuschel Unio crassus (PHILIPSSON 1788) im Seegraben Kanton Schaffhausen. Mitt. Natf. Ges. Schaffhausen 46: 85–100.
Studer, S., 1789. Faunula Helvetica. Class VI. Vermes. Ordo III. Testacea. - In: W. Coxe, Travels in Switzerland, in a series of letters to William Melmoth, Esq., Volume III: 384–392. London (T. Cadell in the Strand)
Studer, S., 1820. Kurzes Verzeichnis der bis jetzt in unserm Vaterlande entdeckten Conchylien. Naturwissenschaftlicher Anzeiger der Allgemeinen Schweizerischen Gesellschaft für die Gesammten Naturwissenschaften, 3 (11): 83–90; (12): 91–94. Bern [1 May; 1 June]. [reprinted as a separate work under the title: Systematisches Verzeichniss der bis jetzt bekannt gewordenen Schweizer-Conchylien. Bern (Stämpfli: 32 pp. > 5 April)]
Weigand, A. M., A. Jochum, R. Slapnik, J. Schnitzler, E. Zarza & A. Klussmann-Kolb, 2013. Evolution of microgastropods (Ellobioidea, Carychiidae): integrating taxonomic, phylogenetic and evolutionary hypotheses. BMC Evolutionary Biology 13: 18.
Whiting, M. F., 2002. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoologica Scripta 31: 93–104.
Xia, X., 2017. DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. Journal of Heredity 108: 431–437.
Xia, X., 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Molecular Biology and Evolution 35: 1550–1552.
Xia, X., Z. Xie, M. Salemi, L. Chen & Y. Wang, 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution 7: 158.
Yusseppone, M. S., F. Márquez, C. M. Luquet, T. Brey, M. C. Ríos de Molina & I. Rocchetta, 2018. Does shell shape variation play a role in conservation of the long-lived freshwater bivalve Diplodon chilensis (Bivalvia, Hyriidae)? Ecohydrology 11: e1931.
Zieritz, A. & D. C. Aldridge, 2009. Identification of ecophenotypic trends within three European freshwater mussel species (Bivalvia: Unionoida) using traditional and modern morphometric techniques: ECOPHENOTYPIC TRENDS IN FRESHWATER MUSSELS. Biological Journal of the Linnean Society 98: 814–825.
Zieritz, A., J. I. Hoffman, W. Amos & D. C. Aldridge, 2010. Phenotypic plasticity and genetic isolation-by-distance in the freshwater mussel Unio pictorum (Mollusca: Unionoida). Evolutionary Ecology 24: 923–938.
Acknowledgements
We are very grateful to Dr. Francis Cordillot (formerly BAFU) for his generous support at the beginning of the project. Our thanks go to Dr Karl-Otto Nagel (SMF) for supplying additional specimens for research. We are deeply grateful to MSc. Estée Bochud, NMBE, for her helpful comments in image processing and for creating the distribution map. Furthermore, we are very grateful to Dr. Adrienne Jochum for her technical and intellectual support. We are thankful for Dr. Beatrice Vögeli and Isabelle Flöss (Fachstelle Naturschutz Kanton Zürich) for the information concerning U. crassus in the Canton of Zürich. Special thanks goes to the team of the UBELIX cluster of Bern for their amazing, friendly support and troubleshooting. This project was financed by the BAFU under the contract No. 16.0100.PJ/R354-0347.
Funding
Open access funding provided by University of Bern. This project was financed by the BAFU under the contract No. 16.0100.PJ/R354-0347.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
No conflicts to report.
Additional information
Handling Editor: Manuel Lopes-Lima
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pfarrer, B., Carlevaro, A., Stucki, P. et al. New phylogenetic insights on some species of Unionidae from Switzerland (Bivalvia, Palaeoheterodonta, Unionidae). Hydrobiologia 849, 2967–2981 (2022). https://doi.org/10.1007/s10750-022-04907-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-022-04907-8