Abstract
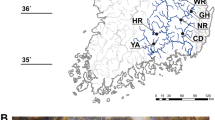
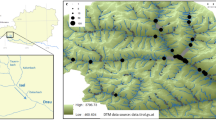
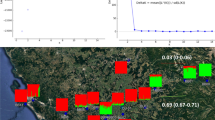
Like in many other parts of the world, Phragmites australis distribution and abundance are changing in Lithuania because of the impact of human activities on aquatic habitats. We studied P. australis genetic diversity patterns and the effect of hydrographic modifications, introduced in the mid-late twentieth century with great impact on Lithuanian landscape. The genetic diversity was studied using chloroplast DNA sequences and nuclear microsatellite markers in natural and straightened river stretches for water regulation. We found haplotypes M and L and their variants in the studied populations. The analysis of microsatellites revealed high genetic diversity within populations and significant structure both at the population and river level. We did not find differences in the distribution of genetic diversity between populations growing in natural and in straightened river stretches; however, at the local level, 5 populations in straightened river sites had higher genetic diversity values than populations in nearby natural sites within the same river, confirming seed establishment in disturbed habitats. Our results demonstrate that anthropogenic river modifications have had an impact on the genetic diversity of P. australis populations; however, disturbance is not the only factor that affects genetic diversity recruitment and dynamics in new P. australis stands.




Similar content being viewed by others
References
Anderson, N. O., L. Jocienė, E. Krokaitė, T. Rekašius, A. Paulauskas & E. Kupčinskienė, 2018. Genetic diversity of Phalaris arundinacea populations in relation to river regulation in the Merkys basin, Lithuania. River Research and Applications 34: 300–309.
Belzile, F., J. Labbé, M. C. LeBlanc & C. Lavoie, 2010. Seeds contribute strongly to the spread of the invasive genotype of the common reed (Phragmites australis). Biological Invasions 12: 2243–2250.
Burnside, N. G., C. B. Joyce, E. Puurmann & D. M. Scott, 2007. Use of vegetation classification and plant indicators to assess grazing abandonment in Estonian coastal wetlands. Journal of Vegetation Science 18: 645–654.
Clevering, O. A. & J. Lissner, 1999. Taxonomy, chromosome numbers, clonal diversity and population dynamics of Phragmites australis. Aquatic Botany 64: 185–208.
Coppi, A., L. Lastrucci, D. Cappelletti, M. Cerri, F. Ferranti, V. Ferri, B. Foggi, D. Gigante, R. Venanzoni, D. Viciani, R. Selvaggi & L. Reale, 2018. AFLP approach reveals variability in Phragmites australis: implications for its die-back and evidence for genotoxic effects. Frontiers in Plant Science 9: 386. https://doi.org/10.3389/fpls.2018.00386.
Dorken, M. E. & C. G. Eckert, 2001. Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). Journal of Ecology 89: 339–350.
Eller, F., H. Skálová, J. S. Caplan, G. P. Bhattarai, M. K. Burger, J. T. Cronin, W.-Y. Guo, X. Guo, E. L. G. Hazelton, K. M. Kettenring, C. Lambertini, M. K. McCormick, L. A. Meyerson, T. J. Mozdzer, P. Pyšek, B. K. Sorrell, D. F. Whigham & H. Brix, 2017. Cosmopolitan species as models for ecophysiological responses to global change: the common reed Phragmites australis. Frontiers in Plant Science 8: 1–24.
Engloner, A. I. & Á. Major, 2011. Clonal diversity of Phragmites australis propagating along water depth gradient. Aquatic Botany 94: 172–176.
Engloner, A. I. & D. Szegő, 2016. Genetic diversity of riverine reed stands indicating the water regime of the habitat. Ecological Indicators 61: 846–849.
Engloner, A. I., Á. Major & J. Podani, 2010. Clonal diversity along a water depth gradient in a declining reed stand as detected by three different genetic methods. Aquatic Botany 92: 1–8.
Eriksson, O., 1989. Seedling dynamics and life histories in clonal plants. Oikos 55: 231–238.
Erős, T., L. Kuehne, A. Dolezsai, N. Sommerwerk & C. Wolter, 2019. A systematic review of assessment and conservation management in large floodplain rivers: actions postponed. Ecological Indicators 98: 453–461.
Evanno, G., S. Regnault & J. Goudet, 2005. Detecting the number of clusters of individuals using the software structure. A simulation study. Molecular Ecology 14: 2611–2620.
Falush, D., M. Stephens & J. K. Pritchard, 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7: 574–578.
Fér, T. & Z. Hroudová, 2009. Genetic diversity and dispersal of Phragmites australis in a small river system. Aquatic Botany 90: 165–171.
Fuller, M. R., M. W. Doyle & D. L. Strayer, 2015. Causes and consequences of habitat fragmentation in river networks. Annals of the New York Academy of Sciences 1355: 31–51.
Gailiušis, B., J. Jablonskis & M. Kovalenkovienė, 2001. Lithuanian rivers. Hydrography and runoff. Lithuanian Energy Institute, Kaunas.
Guo, W., R. Wang, S. Zhuo, S. Zhang & Z. Zhang, 2003. Genetic diversity and clonal structure of Phragmites australis in the Yellow River delta of China. Biochemical Systematics and Ecology 31: 1093–1109.
Hazelton, E. L. G., M. K. McCormick, M. Sievers, K. M. Kettenring & D. F. Whigham, 2015. Stand age is associated with clonal diversity, but not vigor, community structure, or insect herbivory in Chesapeake Bay Phragmites australis. Wetlands 35: 877–888.
Jablonskis, J., M. Kovalenkovienė & A. Tamkevičienė, 2007. Channel network of the Lithuanian rivers and small streams. Annales Geographicae 40(1): 46–56.
Jacquemyn, H., O. Honnay, K. V. Looy & P. Breyne, 2006. Spatiotemporal structure of genetic variation of a spreading plant metapopulation on dynamic riverbanks along the Meuse River. Heredity 96: 471–478.
Kettenring, K. M., M. K. McCormick, H. M. Baron & D. F. Whigham, 2010. Phragmites australis (common reed) invasion in the Rhode River subestuary of the Chesapeake Bay: disentangling the effects of foliar nutrients, genetic diversity, patch size, and seed viability. Estuaries and Coasts 33: 118–126.
Kettenring, K. M., M. K. McCormick, H. M. Baron & D. F. Whigham, 2011. Mechanisms of Phragmites australis invasion: feedbacks among genetic diversity, nutrients, and sexual reproduction. Journal of Applied Ecology 48: 1305–1313.
Kirk, H., J. Paul, J. Straka & J. R. Freeland, 2011. Long distance dispersal and genetic diversity are implicated in the invasive spread of the common reed Phragmites australis (Poaceae) in Eastern North America. American Journal of Botany 98(7): 1180–1190.
Koppitz, H., 1999. Analysis of genetic diversity among selected populations of Phragmites australis world-wide. Aquatic Botany 64: 209–221.
Koppitz, H. & H. Kühl, 2000. To the importance of genetic diversity of Phragmites australis in the development of reed stands. Wetlands Ecology and Management 8: 403–414.
Koppitz, H., H. Kühl, K. Hesse & J.-G. Kohl, 1997. Some aspects of the importance of genetic diversity in Phragmites australis (Cav.) Trin. ex Steudel for the development of reed stands. Botanica Acta 110: 217–223.
Krokaitė, E., D. Shakeneva, E. Juškaitytė, T. Rekašius, J. Nemaniūtė-Gužienė, J. Butkuvienė, J. Patamsytė, V. Rančelienė, R. Vyšniauskienė, L. Duchovskienė, L. Jocienė, Z. Sinkevičienė, D. Naugžemys, V. Kleizaitė, D. Chmura, N. O. Anderson, D. Žvingila & E. Kupčinskienė, 2019. Nitrogen concentration of the aquatic plant species in relation to land cover type and other variables of the environment. Zemdirbyste-Agriculture 106(3): 203–212.
Kumar, S., G. Stecher, M. Li, C. Knyaz & K. Tamura, 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549.
Lambertini, C., 2016. Heteroplasmy due to chloroplast paternal leakage: another insight into Phragmites haplotypic diversity in North America. Biological Invasions 18: 2443–2455.
Lambertini, C., M. H. G. Gustafsson, J. Frydenberg, M. Speranza & H. Brix, 2008. Genetic diversity patterns in Phragmites australis at the population, regional and continental scales. Aquatic Botany 88: 160–170.
Lambertini, C., B. K. Sorrell, T. Riis, B. Olesen & H. Brix, 2012. Exploring the borders of European Phragmites within a cosmopolitan genus. AoB PLANTS 2012: pls020.
Lambertini, C., W. Y. Guo, S. Ye, F. Eller, X. Guo, X. Z. Li, B. K. Sorrell, M. Speranza & H. Brix, 2020. Phylogenetic diversity shapes salt tolerance in Phragmites australis estuarine populations in East China. Scientific Reports 10: 17645.
Liu, L., M. Yin, X. Guo, J. Wang, Y. Cai, C. Wang, X. Yu, N. Du, H. Brix, F. Eller, C. Lambertini & W. Guo, 2020. Cryptic lineages and potential introgression in a mixed-loidy species (Phragmites australis) across temperate China. Journal of Systematics and Evolution. https://doi.org/10.1111/jse.12672.
Liu, L., J. Wang, X. Ma, M. Li, X. Guo, M. Yin, Y. Cai, X. Yu, N. Du, R. Wang & W. Guo, 2021. Impacts of the Yellow River and Qingtongxia dams on genetic diversity of Phragmites australis in Ningxia Plain, China. Aquatic Botany 169: 103341.
McCormick, M. K., D. F. Whigham, J. R. Stapp, E. L. G. Hazelton, E. K. McFarland & K. M. Kettenring, 2020. Shoreline modification affects recruitment of invasive Phragmites australis. Wetlands Ecology and Management 28: 909–919.
McCormick, M. K., K. M. Kettenring, H. M. Baron & D. F. Whigham, 2010a. Spread of invasive Phragmites australis in estuaries with differing degrees of development: genetic patterns, Allee effects and interpretation. Journal of Ecology 98: 1369–1378.
McCormick, M. K., K. M. Kettenring, H. M. Baron & D. F. Whigham, 2010b. Extent and reproductive mechanisms of Phragmites australis spread in brackish wetlands in Chesapeake Bay, Maryland (USA). Wetlands 30: 67–74.
Meyerson, A. L., J. T. Cronin & P. Pyšek, 2016. Phragmites australis as a model organism for studying plant invasions. Biological Invasions 18: 2421–2431.
Nilsson, C., R. L. Brown, R. Jansson & D. M. Merritt, 2010. The role of hydrochory in structuring riparian and wetland vegetation. Biological Reviews of the Cambridge Philosophical Society 85: 837–858.
Patamsytė, J., D. Naugžemys, T. Čėsnienė, V. Kleizaitė, O. N. Demina, S. I. Mikhailova, V. A. Agafonov & D. Žvingila, 2018. Evaluation and comparison of the genetic structure of Bunias orientalis populations in their native range and two non-native ranges. Plant Ecology 219: 101–114.
Paukštys, B., 2011. Ats. Red. Lietuvos vandens telkinių būklė ir ūkinės veiklos poveikis. Aplinkos apsaugos agentūra prie LR aplinkos ministerijos 631: 467–469.
Peakall, R. & P. E. Smouse, 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28: 2537–2539.
Prach, K. & P. Pyšek, 2001. Using spontaneous succession for restoration of human-disturbed habitats: experience from Central Europe. Ecological Engineering 17: 55–62.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Pyšek, P., H. Skálová, J. Čuda, W.-Y. Guo, J. Suda, J. Doležal, O. Kauzál, C. Lambertini, M. Lučanová, T. Mandáková, L. Moravcová, K. Pyšková, H. Brix & L. A. Meyerson, 2018. Small genome separates native and invasive populations in an ecologically important cosmopolitan grass. Ecology 99: 79–90.
Rosenberg, N. A., 2004. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes 4(1): 137–138.
Saltonstall, K., 2002. Cryptic invasion by a non-native genotype of Phragmites australis into North America. Proceedings of the National Academy of Sciences of the USA 99: 2445–2449.
Saltonstall, K., 2003. Microsatellite variation within and among North American lineages of Phragmites australis. Molecular Ecology 12: 1689–1702.
Saltonstall, K., 2016. The naming of Phragmites haplotypes. Biological Invasions 18: 2433–2441.
Sampson, J. F., M. Hankinson, S. McArthur, S. Tapper, M. Langley, N. Gibson, C. Yates & M. Byrne, 2015. Long-term ‘islands’ in the landscape: low gene flow, effective population size and genetic divergence in the shrub Hakea oldfieldii (Proteaceae). Botanical Journal of the Linnean Society 179: 319–334.
Stabile, J., D. Lipus, L. Maceda, M. Maltz, N. Roy & I. Wirgin, 2016. Microsatellite DNA analysis of spatial and temporal population structuring of Phragmites australis along the Hudson River Estuary. Biological Invasions 18: 2517–2529.
Van de Peer, Y. & R. De Wachter, 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Computer Applications in the Biosciences 10: 569–570.
Vekemans, X., T. Beauwens, M. Lemaire & I. Roldán-Ruiz, 2002. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology 11: 139–151.
Yeh, F.C., R. Yang & T. Boyle, 1999. POPGENE: Microsoft Windows-based freeware for population genetic analysis. Release 1.31, University of Alberta, Edmonton.
Žikulinas, J. & A. Česnulevičius, 2009. Hidrografinio tinklo pokyčiai Lietuvos mažųjų upių baseinuose. Geografija 45(1): 62–68.
Funding
This work was supported by the Research Council of Lithuania (SIT-2/2015).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guest editors: Franziska Eller, Hans Brix, Brian K. Sorrell & Carlos A. Arias / Wetland ecosystems: functions and use in a changing climate
Rights and permissions
About this article
Cite this article
Naugžemys, D., Lambertini, C., Patamsytė, J. et al. Genetic diversity patterns in Phragmites australis populations in straightened and in natural river sites in Lithuania. Hydrobiologia 848, 3317–3330 (2021). https://doi.org/10.1007/s10750-021-04606-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-021-04606-w