Abstract
Prior to burning down in 1906 CE, Point Alones in the Monterey Bay region of Central California was home to one of the largest Chinese fishing communities in the United States of America. Both historical records and the recovery of numerous cartilaginous fish (Chondrichthyes) vertebrae during archaeological excavations of the village indicate sharks were among the taxonomic groups being regularly harvested by its inhabitants. However, as shark vertebrae are difficult to identify past the family-level using conventional morphology-based approaches, our understanding of the Point Alones shark fishery remains incomplete. In this study, we address this issue by using ancient DNA analysis to assign species-level identifications to a sample of 54 shark vertebrae from the site. We successfully amplified a 173 bp fragment of the mitochondrial cytochrome c oxidase I gene from 47 of the 54 analyzed specimens (87.03%). Our results indicate that Tope Shark (Galeorhinus galeus; n = 39) was the primary focus of the site’s shark fishery, with Brown Smooth-Hound (Mustelus henlei; n = 7) and Leopard Shark (Triakis semifasciata; n = 1) also harvested to a lesser extent. All three of these species are found locally in the waters overlying the continental shelf, suggesting Chinese fishers were harvesting sharks from these coastal environments. While some of the sharks caught by fishers from Point Alones were likely being consumed at the village, historical records suggest a significant number of fins from harvested Tope Sharks were also likely being exported to China and other diaspora communities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
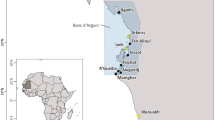
The waters in and around Central California’s Monterey Bay support a diverse community of fishes, with 507 species known to currently or historically inhabit this region (Burton & Lea, 2019). Drawn in part by the region’s marine biodiversity, Chinese immigrants, primarily from southern China, established fishing villages at Point Alones (Fig. 1) and other areas around Monterey Bay in the mid-nineteenth century (Armentrout-Ma, 1981; Lydon, 1985). Historical records and zooarchaeological data indicate Chinese fishers from Point Alones and other settlements took advantage of Monterey Bay’s rich resources to catch a tremendous range of marine invertebrates and fish species (Armentrout-Ma, 1981; Lydon, 1985). Archaeological excavations at Point Alones have recovered numerous ossified shark vertebrae, indicating that sharks were among the taxa regularly harvested by fishers operating from the village. The existence of a Chinese diaspora shark fishery in the region is also attested to in historical records (Lydon, 1985).
Map showing the location of the Point Alones Chinese fishing village and other key geographic features mentioned in the text. The map was created with QGIS version 3.32 (http://www.qgis.org) using a base map from ESRI (https://www.esri.com)
In California, landing records for individual shark species are not available prior to the 1940s (Free et al., 2022; Ripley, 1946). However, historical records indicate that most of the sharks landed in California during the mid-twentieth century were Tope Sharks (Galeorhinus galeus) (Byers, 1940; Ripley, 1946; Walford, 1931). During the nineteenth century, in addition to Tope Sharks, Chinese fishers operating in California were also known to harvest Shovelnose Guitarfish (Pseudobatos productus), Gray Smooth-Hound (Mustelus californicus), and Brown Smooth-Hound (Mustelus henlei) (Jordan, 1905; Jordan & Gilbert, 1882). Nonetheless, due to the lack of species-specific landing records, more precise information about the relative importance of these species to Chinese operated fisheries in the Monterey Bay region is unavailable. It is also unknown whether other shark species were harvested by these fisheries. Documenting when commercial fisheries for particular shark species began in Monterey Bay and the intensity of these fisheries can contribute to the present-day management of shark populations in the region. Without information about the exploitation history of species and when fisheries began to impact populations, it is difficult to establish conservation baselines representative of conditions prior to fishing-induced changes (Andrews et al., 2022; Barrett, 2019; Jackson et al., 2001; Pauly, 1995).
In the absence of historical catch records, zooarchaeological data can be used to reconstruct past fisheries (e.g., Barrett et al., 2004; Guiry et al., 2021; Hawkins et al., 2019; Kennedy, 2017; Lambrides et al., 2019; McKechnie & Moss, 2016). However, multiple factors hamper efforts to document past harvests and uses of cartilaginous fish (Chondrichthyes; shark, rays, skates, sawfishes, and chimaeras) using zooarchaeological data. Due to the cartilaginous nature of their skeletons, Chrondrichthyes remains are often not preserved in large quantities at archaeological sites, leading to an underestimation of their economic importance (Rick et al., 2002). The range of cartilaginous fish elements that tend to preserve archaeologically is also limited, which poses interpretative issues. Of the six skeletal elements from cartilaginous fish that are commonly recovered from archaeological contexts (vertebrae, teeth, fin ray spines, tail spines, dermal denticles, and rostra), only teeth can be readily identified to the species-level using conventional morphology-based zooarchaeological approaches (Rick et al., 2002). However, as is the case at Point Alones, many archaeological cartilaginous fish assemblages lack teeth and consist entirely of ossified vertebrae. Since the vertebral morphology of many closely related taxa is similar, reliably identifying cartilaginous fish vertebrae to the species-level through conventional zooarchaeological methods is often challenging or impossible (Gobalet et al., 2004; Rick et al., 2002; Shepherd & Campbell, 2021). Moreover, significant intra-specific variation in vertebrae morphology related to ontogeny and spinal column position has been observed among cartilaginous fishes, further hindering the identification of these elements to the species-level (Rick et al., 2002; Shepherd & Campbell, 2021). These barriers to species identification are compounded by the fact that many zooarchaeological reference collections lack comprehensive collections of cartilaginous fish species (Shepherd & Campbell, 2021). Given these issues, cartilaginous fish remains are typically identified to the family-level or higher, remain completely unidentified, and/or in some cases may be misidentified or assigned different identifications by different analysts (Gobalet, 2001; Gobalet et al., 2004; Rick et al., 2002; Shepherd & Campbell, 2021). Comparisons of morphology- and DNA-based identifications of cartilaginous fish vertebrae by Shepherd and Campbell (2021) found that most of their morphology-based identifications were incorrect, with confamiliar misidentifications being particularly common.
As DNA variation exists among species, ancient DNA (aDNA) analysis can often be used to assign species identifications to archaeological fish remains that lack diagnostic morphological features (Yang et al., 2004). This approach to species identification involves comparing sequences obtained from archaeological specimens for one or more genetic markers to reference sequences whose taxonomic identity is known (Yang et al., 2004). Such DNA-based approaches have been successfully used to identify cartilaginous fish remains recovered from sites in various locales including Madagascar (Douglass et al., 2018; Grealy et al., 2016), New Zealand (Seersholm et al., 2018), and California (Palmer et al., 2018). In our study we identify the shark species harvested by the Chinese inhabitants of Point Alones and evaluate their relative importance by applying aDNA analysis to a sample of 54 shark remains. We also sought to use the ecology of the identified species to reconstruct the fishing strategies employed by the Point Alones shark fishery.
Archaeological and Historical Context
Point Alones was located on the southern shore of Monterey Bay, California, on the Monterey Peninsula in the present-day city of Pacific Grove (Fig. 1). The site was home to a large Chinese community, primarily fishers and their families, from the late 1850s through the turn-of-the-century (Lydon, 1985). Unlike many Chinese diaspora communities in the United States, which were often predominately composed of men, a significant proportion of Point Alones’ population were women (43% in 1870) and children (32% in 1870) (Lydon, 1985). In addition to residences and fishing-related facilities, such as large drying racks (Collins, 1892; Fig. 2), the village also had an employment agency, stores, and various social and religious amenities (e.g., cemetery, shrine) (Lydon, 1985; Williams, 2011). In 1906, the village was abandoned after it burned down in a fire that is thought by some to have been deliberately set with the intention of driving the Chinese community out (Lydon, 1985). While Point Alones is characterized as a Chinese settlement, it is important to note that Chinese diaspora communities were often multiethnic and multiracial spaces (Fong et al., 2022). In the case of Point Alones, both Japanese fisherman (Lydon, 1997) and an African American woman (Williams, 2011) are known to have resided in the village.
Drawing from Collins (1892) of Chinese fishers at Point Alones curing a large quantity of squid on drying racks, ca. 1888–1889. Digitized image courtesy of the University of Washington Libraries Freshwater Marine Image Bank
Most Point Alones residents made their livelihoods harvesting, processing, and exporting a diverse array of marine resources. Sharks, squid, finfish, abalone, kelp, sea mammals, and salt are all recorded among the resources exploited by the community (Armentrout-Ma, 1981; Lydon, 1985). Historical newspapers and government reports describe Chinese individuals fishing from both the shore and watercraft and employing a wide range of methods, including trolling, set lining, gillnetting, bag netting, seining, hand fishing, and the use of fish traps (Jordan, 1887a; Collins, 1892; Daily Alta California, 1887). Men, women, and children all played an active role in the village’s fishing industry (Lee, 2006; Lydon, 1985). Although most fishers were men, women from the community also fished from time to time, in addition to repairing fishing gear, developing new processing and preservation methods, and together with the children and men processing and drying fish and other marine resources (Lee, 2006; Lydon, 1985). Both the large amount of seafood exported from Point Alones and the plethora of seafood products produced there were highlighted by famed fish taxonomist Jordan (1887a, b, 1892), who visited and described the village on multiple occasions. Fish specimens collected by Quock (Kwok) Tuck Lee and other fishers from Point Alones were fundamental to many of the zoological studies carried out by Jordan and other researchers associated with Stanford University’s neighbouring Hopkins Marine Station (originally known as the Hopkins Seaside Laboratory) (Kohrs, 2013).
Williams’ (2011) archaeological excavations at Point Alones (Site number: CA-MNT-104) uncovered several refuse lenses and midden deposits rich in faunal remains. The associated fish assemblage has been the subject of ongoing zooarchaeological analysis by Kennedy, and preliminary results reinforce historical accounts indicating Chinese fishers were making use of a wide range of marine resources. Although the faunal assemblage is dominated by rockfishes (Sebastes spp.) and flatfishes (Pleuronectidae), dozens of other taxa, including Wolf Eels (Anarrhichthys ocellatus), Pacific Jack Mackerel (Trachurus symmetricus), Pacific Hake (Merluccius productus), surfperches (Embiotocidae), and houndsharks (Triakidae), appear in nearly all contexts at the site and clearly formed important components of village fisheries. In the context of broader Chinese diaspora zooarchaeology, the identified houndshark vertebrae are notable as shark vertebrae of any kind are rarely recovered from Chinese consumer sites in the western United States. This is likely due to the trade of shark fins and tails rather than meat or whole dried animals to consumer sites, making the analysis of the shark assemblage from Point Alones a rare opportunity to better understand the specifics of Chinese-operated shark fisheries.
Materials and Methods
Decontamination and DNA Extraction
A total of 54 shark vertebrae from the fish assemblage collected by Williams (2011) were selected for aDNA analysis (Fig. 3). The analyzed specimens originated from various excavation contexts and were all morphologically identified as houndsharks (see Supplementary Table 1 for details). Decontamination, DNA extraction, and PCR setup procedures were performed in a positively pressured laboratory with a UV-HEPA air filtration system in the Department of Archaeology, Simon Fraser University (Burnaby, BC, Canada), that is dedicated to the analysis of aDNA (Cooper & Poinar, 2000; Yang & Watt, 2005). Following Speller and colleagues (2012), all the specimens were decontaminated prior to DNA extraction using a combination of bleach and UV light. The specimens were submerged in a 100% commercial bleach solution (7.55% w/v NaOCl; Clorox Company) for 6–8 min; submerged in distilled water for 30 s–2 min; submerged again in distilled water for 6–10 min; and UV irradiated in a crosslinker for 30 min (15 min on two sides). DNA was extracted from the decontaminated samples using a modified silica spin-column method (Yang et al., 1998, 2008). Following decontamination, the samples were incubated in a rotating hybridization oven at 50 °C overnight in 4.5 mL of lysis buffer (0.5 M EDTA [pH 8.0], 0.25% SDS, and 0.5 mg/ml proteinase K). The samples were then centrifuged and 3–4 mL of the resulting supernatant was concentrated to approximately 100 µL using Amicon 10 kDA centrifugal filter units (Millapore). The samples were then purified with a QIAquick PCR Purification Kit (Qiagen) following the manufacturer’s directions, with a final elution volume of 100 µL. The analyzed specimens were processed in batches that consisted of up to seven specimens. Each batch included a blank extraction control that was subjected to amplification in order to dectect instances of contamination (Cooper & Poinar, 2000). To further ensure our results were not due to contamination, DNA extraction, amplification, and sequencing in both directions was repeated for nine samples (SHK16, SHK18, SHK19, SHK26, SHK30, SHK31, SHK35, SHK47, and SHK54) (Cooper & Poinar, 2000).
PCR Amplification and Sequencing
To identify the specimens, we amplified and sequenced an approximately 173 bp fragment (approximately 127 bp without the primer sequences) of the DNA barcode or Folmer region (Folmer et al., 1994; Hebert et al., 2003) of the mitochondrial cytochrome c oxidase I (COI) gene spanning positions 5479 to 5651 of the Tope Shark mitochondrial genome (GenBank accession number: ON652874; Winn et al., 2024). We amplified this region with two published sets of universal primers for cartilaginous fish (Fields et al., 2015; Shepherd & Campbell, 2021). These primer sets consist of a single shared reverse primer (Shark COI-MINIR) designed by Fields et al. (2015) and one of two forward primers (FishF1and FishF2) designed by Ward et al. (2005) (Table 1). Previous studies have demonstrated that these primer sets can amplify the targeted COI fragment from a broad range of shark species and that this fragment can discriminate many cartilaginous fish species (Fields et al., 2015; Shepherd & Campbell, 2021). Furthermore, the successful application of these primers to archaeological cartilaginous fish remains by Shepherd and Campbell (2021) demonstrate that they are sufficiently sensitive to amplify DNA from ancient materials with degraded DNA. For this study, we initially attempted to amplify the targeted COI fragment with primers FISHF1and Shark COI-MINIR. In instances where this primer pair failed to amplify DNA, we attempted to amplify this COI fragment with primers FishF2 and Shark COI-MINIR.
All PCR amplifications and procedures involving amplified DNA were carried out in a negatively pressured post-PCR laboratory located in a different building than the aDNA laboratory (Cooper & Poinar, 2000; Yang & Watt, 2005). PCR amplifications were performed on a Mastercycler Personal or Gradient thermal cycler (Eppendorf) in a 30 µL reaction volume that included 1.5 × PCR Gold Buffer (Applied Biosystems), 2 mM MgCl2, 0.2 mM of each dNTP, 0.3 μM of primer Shark COI-MINIR, 3 μM of primer FishF1or FishF2, 1 mg/mL BSA, 3 μl DNA sample, and 0.75–1 U AmpliTaq Gold (Applied Biosystems). The thermal cycling program comprised of an initial denaturation step at 95 °C for 12 min followed by 60 cycles at 95 °C for 30 s (denaturation), 52–54 °C for 30 s (annealing), and 70 °C for 40 s (extension), and a final extension step at 72 °C for 7 min. A negative control was included in each PCR run to monitor for contamination (Cooper & Poinar, 2000).
Following amplification, 5 μL of PCR product from each specimen was pre-stained with SYBR Green I (Invitrogen), separated by electrophoresis on a 2% agarose gel, and visualized with a Dark Reader transilluminator (Clare Chemical Research). Successfully amplified specimens were directly Sanger sequenced with the forward or reverse amplification primers at Eurofins Genomics. To assess the replicability of our results, amplification and sequencing were repeated at least once for specimens that did not undergo repeat DNA extraction (Cooper & Poinar, 2000; Winters et al., 2011). PCR products generated through repeat amplifications were sequenced in the opposite direction of the initial sequencing reaction.
Species Identification
The obtained sequences were visually examined and edited, trimmed to remove the primer sequences, and compiled into consensus sequences using ChromasPro version 2.1.8 (http://technelysium.com.au/wp/chromaspro). To assign a species-level identification to the analyzed specimens, the obtained consensus sequences were compared to reference sequences accessioned in the GenBank Nucleotide Collection (Sayers et al., 2023) using BLASTn (Altschul et al., 1990) and cross-checked against reference sequences accessioned in the Barcode of Life Data System (BOLD) Public Record Database with the BOLD Identification Engine (Ratnasingham & Hebert, 2007). Following Fields et al. (2015), a species-level identification was assigned to a specimen if both the BLASTn and BOLD Identification Engine results indicated there was a perfect match (100% sequence similarity) with published sequences from one species.
Results
PCR Amplification
A fragment of COI was amplified with the FishF1and Shark COI-MINIR primer pair from 47 of the analyzed 54 specimens (87.03%) (Table 2; Fig. 4). In the case of the 9 specimens where DNA extraction was performed twice (SHK16, SHK18, SHK19, SHK26, SHK30, SHK31, SHK35, SHK47, and SHK54), the targeted COI fragment was successfully amplified from both extractions. This fragment was also successfully amplified at least twice from the remaining 38 specimens with amplifiable DNA that were not subject to repeat DNA extraction. Use of the FishF2 and Shark COI-MINIR primer pair did not result in the amplification of DNA from any additional specimens. Two of the specimens (SHK23 and SHK24) that did not yield DNA were calcined as a result of burning and the failure to amplify DNA from these specimens likely reflects heat-induced DNA degradation (Supplementary Table 1; Emery et al., 2022). None of the specimens from which DNA was successfully amplified exhibited signs of burning (Table 2; Supplementary Table 1). DNA was amplified from one of the blank extraction controls. However, sequence analyses indicate that the DNA amplified from this blank extraction control is human in origin. No DNA was amplified from any of the other blank extraction controls or negative PCR controls.
Negative image of electrophoresis gel of PCR products obtained with primers FishF1and Shark COI-MINIR from a subset of the archaeological shark vertebrate specimens from Point Alones (SHK#). BK denotes the blank extraction control that was processed alongside the specimens and NEG denotes the negative PCR control that was included in the PCR run. The 100 bp ladder is from Invitrogen. An unprocessed image of the electrophoresis gels is presented in the supplementary material (Supplementary Fig. 1)
Species Identification
The COI sequences obtained from all 47 of the specimens with amplifiable DNA matched reference sequences from sharks accessioned in GenBank and BOLD (Table 2). Sequences obtained from different extractions and/or amplifications conducted on the same specimen were concordant. All 47 of the COI sequences are 127 bp long and have been submitted to GenBank under accession numbers OR731536 through OR731582. Using the criteria outlined above, species-level identifications could be assigned to 8 (17.02%) of the 47 specimens (Table 2). The majority of remains (n = 39, 82.98%) initially could not be identified to the species-level (Table 2). BLASTn searches of these specimens’ COI sequences against GenBank indicated they were identical to sequences from both Tope Shark and an unidentified ground shark (Carcharhiniformes; GenBank accession number: GU805891) (Table 2). However, an examination of the BOLD record for this unidentified ground shark (BOLD sequence ID: ELAME541-09) revealed that this specimen was a Tope Shark. If this specimen is treated as a Tope Shark, all of the remaining 39 specimens (82.98%) with amplifiable DNA could be assigned to the species-level. In total, 39 (82.98%) of the specimens were identified as Tope Shark, 7 (14.89%) were identified as Brown Smooth-Hound, and 1 (2.13%) was identified as Leopard Shark (Table 2). These identifications are consistent with the morphology-based identification of these sharks as houndsharks, as all three of these species belong to this family.
Discussion
Authenticity of the Ancient DNA Results
Upon an organism’s death, the mechanisms that repaired its DNA while it was living cease, exposing their DNA to degradative processes that reduce the quantity and quality of DNA preserved within their remains (Dabney et al., 2013). As a result of this degradation, archaeological materials are susceptible to contamination with modern DNA and carryover contamination from previous amplifications (Cooper & Poinar, 2000; Yang & Watt, 2005). While the occurrence of contamination can never be entirely ruled out, the balance of probabilities suggest our a DNA results are authentic. All pre-PCR procedures in this study were conducted in a positively pressured dedicated aDNA laboratory physically separated from the negatively pressured post-PCR facility (Cooper & Poinar, 2000; Yang & Watt, 2005). All personnel working in the aDNA laboratory wore protective clothing (gloves, masks, and coveralls) and there was a one-way flow of goods and personnel from the aDNA to post-PCR laboratory (Yang & Watt, 2005). In addition, all of the remains were decontaminated prior to DNA extraction through a combination of bleach and UV irradiation (Yang & Watt, 2005). While these controls limited the potential for contamination with exogenous DNA, the amplification of human DNA from one of the blank extraction controls indicates there was an occurrence of contamination. Although its precise source cannot be identified at this time, this human DNA might have been shed from individuals working in the aDNA laboratory or represent reagent contamination that occurred at the manufacturing plant (Leonard et al., 2007). Nonetheless, the failure to amplify shark DNA from the blank extraction or negative PCR controls indicates that systematic contamination with modern or previously amplified shark DNA did not occur (Cooper & Poinar, 2000). The authenticity of our aDNA data is further supported by the fact that for each specimen sequences obtained through repeat PCR amplifications on the same or different DNA extracts were in agreement (Cooper & Poinar, 2000). Finally, we identified multiple species, often within the same extraction batch, which reduces the likelihood that contamination can account for our results, as it would need to have originated from multiple sources (Yang et al., 2004).
Fishing Strategies
Observations in the mid-twentieth century indicate that most sharks being landed by fishers operating from California were Tope Shark (Byers, 1940; Ripley, 1946; Walford, 1931). The predominance of Tope Sharks among the archaeological shark remains from Point Alones indicate that by the turn-of-the-century this pattern had already emerged among Chinese fishers in the Monterey Bay region. While the Point Alones shark fishery appears to have specialized in harvesting Tope Sharks, this species was not the only one harvested. Our data indicate that Brown Smooth-Hound and Leopard Shark were also landed by Chinese fishers to a lesser degree. Although both Tope Shark and Brown Smooth-Hound are documented in historical records as having been harvested by Chinese operated fisheries in California (Jordan, 1905; Jordan & Gilbert, 1882), to the best of our knowledge, our identification of Leopard Shark at Point Alones is the first evidence for these fisheries harvesting this species. Given its low frequency, it is also possible the identified Leopard Shark vertebrae was incidentally transported to the site as part of the stomach contents of another fish, such as Tope Shark, which is known to consume sharks (Ripley, 1946).
All three of the identified shark species are present within the Monterey Bay area today (Burton & Lea, 2019), suggesting the Point Alones shark fishery focused on local fishing grounds. Additional support for the local orientation of the shark fishery can be found in historical records. In 1892, Collins (1892) noted that Chinese fishers from Point Alones largely focused their efforts in the coastal waters of Monterey Bay between Point Alones and Cypress Point (Fig. 1). Locally, Tope Sharks, Brown Smooth-Hound, and Leopard Sharks are most commonly encountered in the neritic zone, which encompasses coastal waters overlying the continental shelf (Ebert, 2003). Historically, the majority of Tope Sharks landed by commercial fisheries in the Monterey Bay region were captured at depths between 11 to 20 fathoms (approximately 20.12 to 36.58 m) (Ripley, 1946). However, Tope Sharks are also known to occur in much deeper offshore waters at depths up to 821 m and shallower inshore waters less than 20 m deep (Ebert, 2003; Ripley, 1946). The other two species identified at Point Alones tend to occur in shallower waters relative to Tope Sharks. Brown Smooth-Hound have a depth range that extends from the intertidal to 369 m while Leopard Sharks can be found in waters up to 191 m deep but are most common in waters spanning the intertidal to 20 m (Ebert, 2003; Love et al., 2021). In the context of Monterey Bay, Leopard Sharks are especially common in the waters in and around Elkhorn Slough (Fig. 1), a tidal slough and estuary located in eastern Monterey Bay, though this species also ranges through other habitats in the area (Carlisle & Starr, 2009; Yoklavich et al., 1991). These ecological preferences suggest that efforts of the Chinese shark fishery operating from Point Alones were largely concentrated in the neritic zone.
Within the Monterey Bay region, sampans were the primary watercraft used by Chinese fishers (Lydon, 1985). The use of sampans by fishers at Point Alones is reflected in historical photographs, which often show these vessels hauled up onto the beach in front of the village (Fig. 5) While the term sampan can refer to boats with a diverse range of designs (Van Tilburg, 2007), Chinese-style sampans such as those used at Point Alones are typically relatively small flat-bottomed wooden vessels that lack a keel and are propelled by sail or sculling (Folkard, 1901; Lydon, 1985). The flat-bottomed and keelless nature of sampans would make these vessels unstable in rough waters, like those commonly encountered far offshore, and are, therefore, typically used in inland and coastal waters, such as bays (Folkard, 1901). David Starr Jordan (1887b), for instance, described the sampans operated by Chinese fishers from the nearby village of Pescadero as being “clumsy”. Reliance on these vessels may have limited Chinese fishers’ ability to target locally available shark species more common in the open ocean, resulting in the focus on coastal species that we observed. On southern California’s Channel Islands, Braje and Bentz (2015) suggest sampans’ limited open-ocean capabilities similarly contributed to the development of a logistically organized foraging strategy among Chinese operated abalone fisheries.
In addition to sharing a preference for coastal waters, all three of the shark species we identified are typically found in the benthic zone (Ebert, 2003), suggesting the Point Alones shark fishery may have been primarily targeting benthic environments. Tope Sharks, however, have been observed migrating throughout the water column. In Australia, West and Stevens (2001) observed that while Tope Sharks were found near the bottom during the day, individuals often moved higher up the water column at night. They suggest that this migratory pattern reflects the species pursuing prey, such as cephalopods, that have similar diel movements. Pursuing cephalopods and other prey to the surface at night might have facilitated the capture of Tope Sharks by Chinese fishers. To simultaneously avoid competition with and persecution from European fishers and respond to market pressures in China, Chinese fisheries in the Monterey Bay region began to increasingly focus on harvesting and exporting squid during the late nineteenth century (Armentrout-Ma, 1981; Chiang, 2004; Lydon, 1985). Squid fishing was typically conducted from boats at night by torchlight, which attracted squid that could then be seined en masse (Collins, 1892). Historical records indicate that this nighttime squid fishing was conducted “during moonless nights” (Wave, 1897), which roughly corresponds with when Tope Sharks most frequently ascend the water column (West & Stevens, 2001). Squid fishing also primarily occurred in spring and early summer when large numbers of squid moved inshore to spawn (Bentz & Braje, 2020; Collins, 1892). This period overlaps with spring to summer spawning period of Tope Sharks that sees large numbers of individuals congregating in shallow waters (Jordan & Gilbert, 1882). It is possible that Tope Sharks pursuing prey into shallower waters at night were attracted to these congregating squids, allowing them to be co-harvested alongside squid. Stomach content analysis of Tope Sharks from California by Ripley (1946) indicate squid were indeed historically a common prey item for this species. By leveraging predator-prey relations to harvest multiple taxa at once, such ‘prey as bait’ fishing strategies can help increase the overall productivity of a fishery (Monks, 1987).
A Shark Fin Fishery?
Chinese diaspora and home (qiaoxiang) communities were linked through business, kin, clan, and other personal relationships to other Chinese communities across the Pacific World (Hsu, 2006; Kennedy & Rose, 2020; Voss et al., 2018). These connections were the basis of a vast multi-sited network that enabled the movement of goods, money, people, postage, and knowledge between China and diaspora communities as well as among diaspora communities (Hsu, 2006; Kennedy et al., 2022; Yu, 2020). This trade was primarily conducted by jinshanzhuang or Gold Mountain firms, which were businesses largely based in Hong Kong that specialized in shipping commodities between China and diaspora communities (Hsu, 2006). Through jinshanzhuang and affiliated merchants, Chinese diaspora communities were able to obtain a wide variety goods from China and other regions, including foodstuffs, books and magazines, ceramics and other kitchenware, clothing, and medicine (Hsu, 2006; Kennedy et al., 2018, 2022; Voss et al., 2018). In addition to providing diaspora consumers with access to goods from Asia, the Chinese diaspora used jinshanzhuang to send letters and remittances to their home communities as well as export locally procured and/or produced goods that were in demand in China (Bentz & Braje, 2020; Hsu, 2006; Kennedy & Rose, 2020). These firms also functioned as de facto immigration consultants, facilitating the migration of individuals from China to North America (Hsu, 2006).
One of the major nodes in this multi-sited trans-Pacific network was the Monterey Bay region. Historical records indicate that Chinese communities in Monterey Bay were exporting a substantial quantity of dried fish and other marine products, including squid, abalone (Haliotis sp.) and kelp (Bentz & Braje, 2020; Lydon, 1985). One estimate indicates that each year about 100 tons of dried fish were being shipped from the Monterey Bay area by the Chinese community (Anonymous, 1875). Dried seafood products from the region were initially transported north by rail or ship to San Francisco from where they were shipped on to China and other diaspora communities (Anonymous, 1875; Collins, 1892). At Point Alones, sharks were among the taxonomic groups being dried and exported. One contemporary observer writing in the Monterey Gazette (1864) noted that “everything from a shark to a shiner is dried, pickled or salted” by the Chinese fishers at Point Alones “preparatory to shipment abroad”. Dried shark fins were a particularly important trade item. The same observer noted that Point Alones was “steadily thriving upon the profits of dried fish, shark fins and abelones [sic]” (Monterey Gazette, 1864).
In China, dried shark fins were and continue to be a luxury ingredient that, after being rehydrated, can be used in a variety of dishes including soup preparations and stir fries or dressed and served on their own (Simoons, 1991). While the consumption of shark fins dates back to at least the Song dynasty (960 to 1279 CE) (Freeman, 1977), demand increased during the Qing dynasty (1644 to 1912 CE) when they became a mainstay of feasts and available as part of multi-course meals served in high-end restaurants (Spence, 1977). This growing demand for shark fins in China, which exceeded local supplies (Simoons, 1991), likely stimulated the development of the specialized Tope Shark fishery at Point Alones. As is the case today (Fields et al., 2015; Vannuccini, 1999), the fins from a number of shark species were historically dried and sold for consumption (Simoons, 1991; Walford, 1931). However, the dorsal and caudal fins of Tope Sharks were highly prized by Chinese fishers in California as their quality was regarded as superior to other species available in the state (Byers, 1940; Jordan, 1905; Jordan & Gilbert, 1882; Walford, 1931). The value attached to the fins of Tope Shark is reflected in one of the common names for the species in North America: Soupfin Shark. Reflecting their high value status, Tope Shark fins commanded a relatively high price, with Jordan (1905) noting that each pair sold for about US$ 1 − 2 (roughly US$ 35 − 70 today). As Tope Shark are absent from China, sourcing dried shark fins from this species represents the localization (sensu Chee-Beng, 2011) of this good's production, mirroring the adaption to varying ecological, social, and legal circumstances observed in other aspects of food consumption and production practices among the Chinese diaspora in the United States (e.g., Bentz & Braje, 2020; Cummings et al., 2014; Kennedy, 2015, 2017; Kennedy & Guiry, 2023; Kennedy et al., 2018; Popper, 2020; Sunseri, 2020).
Although most of the fish landed by Chinese diaspora fisheries in California was being exported to China (Collins, 1892), a portion of the dried shark fins produced at Point Alones from Tope Sharks were also likely being sold to Chinese communities in western North America. Contemporary accounts from California indicate shark fins were a delicacy that were served at banquets hosted by the local diaspora community and at local restaurants (e.g., Daily Alta California, 1866; Daily Humboldt Times, 1887; Daily Los Angeles Herald, 1900; Evening Sentinel, 1907; Jordan, 1887a, Los Angeles Herald, 1904). For example, in 1866, shark fin with eggs and boiled shark fin were two dishes included in a three-course banquet honouring the heads of the American diplomatic missions to China and Japan—Anson Burlingame and Robert Bruce Van Valkenburg—that was hosted at the Hang Heong Restaurant in San Francisco by Tung Yee and Company, Chy Lung and Company, and Wing Wo Sang and Company (Daily Alta California, 1866). This product would have been economically out of reach for most nineteenth-century Chinese migrants prior to emigrating, but economic opportunities in the United States provided new wealth that allowed for the purchase of previously unaffordable foodstuffs (Kennedy et al., 2018). Today, shark fins continue to be luxury goods, with shark fin soup being served at banquets in celebration of holidays and major life events (Anderson, 1988; Simoons, 1991).
While the Point Alones Tope Shark fishery likely developed in response to the demand for shark fins, it should be noted that the fishery does not appear to have been “finning” sharks. Finning is a practice that involves removing the fins from sharks and discarding their carcasses at sea (Vannuccini, 1999). But the presence of Tope Shark vertebrae at the site suggests entire shark carcasses were being landed by Chinese fishers operating from Point Alones. A historical photograph likewise confirms whole sharks were brought back to Point Alones, where their fins were subsequently removed (Fig. 6). This is not unexpected as the remainder of the sharks had a commercial value. Cuts of Tope Shark were marketed locally as “grayfish, fillet of sole or swordfish (Byers, 1940:23)” to circumvent negative perceptions among Euro-Americans about shark meat (Walford, 1931). Although shark meat is relatively poorly regarded in North America, the firm white flesh of Tope Sharks is considered relatively good (Ebert, 2003; Smith & Susumu, 1979). In addition to the meat, the livers and carcasses of Tope Sharks were also rendered into oil and fish meal (Byers, 1940; Jordan, 1905).
Photograph taken circa 1900 by Alice Iola Hare showing a woman at Point Alones butchering fish that Lydon (1985) has identified as sharks. Note the fins have been removed from the sharks. Photograph courtesy of the Bancroft Library at University of California, Berkeley (Call Number: BANC PIC 1905.05129-PIC)
The other species taken by Point Alones fishers likewise had uses beyond being a source of fins. Like Tope Sharks, Leopard Shark meat is comparatively desirable (Ebert, 2003; Smith & Susumu, 1979), and it is possible Point Alones fishers were similarly selling fillets from this species. On the other hand, Jordan and Gilbert (1882) note that Brown Smooth-Hound was primarily a bait fish and while it is possible the site’s occupants may have been using this species in this manner, it may have also been sold as food. While not a prized foodstuff, Chinese fishers would ship dried and salted young Grey Smooth-Hound (Mustelus californicus)—a closely related species—to Chinese railroad labourers (Jordan & Gilbert, 1882). Beyond these markets and uses, it is likely that Point Alones fishers and their families also ate Tope Shark and other species they caught in the village.
Conclusion and Future Directions
Through the DNA-based species identification of archaeological shark remains, we have presented new insights into the nineteenth- through the early twentieth-century Chinese diaspora shark fishery at Point Alones in the Monterey Bay region of Central California. Historical and zooarchaeological data indicate fishers operating from this settlement harvested a diversity of marine resources. In contrast, our data indicated that, while other species such as Brown Smooth-Hound and Leopard Shark were occasionally caught, the Point Alones shark fishery was highly specialized, focusing largely on Tope Sharks. Tope Shark fins were prized, and the targeting of this species was likely a response to the demand for shark fins in China and to a lesser extent in other diaspora communities. Intriguingly, Tope Sharks are known to ascend the water column to feed on squid, another common export from Point Alones. This behavioral association suggests an ecologically sophisticated ‘prey as bait’ fishing strategy was employed by Point Alones fishers allowing for the convenient harvest of both species simultaneously. Overall, our results showcase how market forces that traversed the Pacific shaped the orientation of Chinese extractive industries in the United Sates, as well as drove unique fishing strategies that reduced conflict with non-Chinese fishers and took advantage of locally available resources. In addition, the ecology of the identified species suggests Point Alones fishers largely focused on the local neritic zone, which possibly reflected the limitations of the traditional inshore Chinese watercraft used by fishers. Our findings also have significant implications for present-day Tope Shark conservation efforts. The existence of an intensive commercial Tope Shark fishery at the turn-of-the-century suggests that populations on the North American Pacific Coast may have been subject to fishing pressures that affected its demography prior to historically documented declines related to overfishing in the 1940s (cf. Andrews et al., 2023; cf. Guiry et al., 2021; Ripley, 1946).
Although less common, shark remains have been reported at other Chinese diaspora sites in North America (e.g., Badenhorst & Ross, 2019; Kennedy, 2017; Porcasi, 2017; Schulz, 2002). It is expected that the DNA-based species identification of remains from others sites will provide additional details into the harvest and use of sharks by historical Chinese diaspora communities. For instance, generating species-level identifications for shark remains from production sites in other areas will shed light on whether a focus on Tope Shark was widespread and the role local ecological conditions played in structuring shark fisheries (e.g., Hawkins et al., 2019; Lambrides et al., 2019; McKechnie & Moss, 2016). In the case of consumer sites, identifying the species present will generate insights into whether the shark taxa consumed differed from those being locally harvested. Moreover, additional population genetic analyses of DNA-identified shark remains from Chinese and non-Chinese sites also has the potential to shed light on species’ demographic history and historical genetic diversity (e.g., Andrews et al., 2021; Johnson et al., 2018; Martínez-García et al., 2021; Speller et al., 2012). Although few palaeogenetic studies focus on cartilaginous fishes, the recovery of DNA from the remains of sharks and other cartilaginous fish in this and other studies (e.g., Douglass et al., 2018; Grealy et al., 2016; Palmer et al., 2018; Seersholm et al., 2018; Shepherd & Campbell, 2021) highlights the potential for such analyses. Lastly, future work combining DNA-based species identification with other biomolecular approaches, such as stable isotope analysis, provides the opportunity to document the past behaviour and ecology of shark species, and how these have changed in response to human activities (e.g., Andrews et al., 2023; Bas et al., 2020; Braje et al., 2017; Elliott Smith et al., 2023; Guiry et al., 2020a, b, c; Miszaniec et al., 2021). By increasing the amount of data that can be obtained from archaeological shark remains, aDNA and other biomolecular analyses have tremendous potential to address questions about human-shark interactions and shark ecology that have remained out of reach for traditional historical and archaeological approaches.
Availability of Data and Materials
All of the generated DNA sequences have been deposited in GenBank under accession numbers OR731536–OR731582. All other relevant data are presented in the main text of the article or the supplementary information.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Anderson, E. N. (1988). The food of China. Yale University Press.
Andrews, A. J., Di Natale, A., Bernal-Casasola, D., Aniceti, V., Onar, V., Oueslati, T., Theodropoulou, T., Morales-Muñiz, A., Cilli, E., & Tinti, F. (2022). Exploitation history of Atlantic bluefin tuna in the eastern Atlantic and Mediterranean—insights from Ancient Bones. ICES Journal of Marine Science, 79(2), 247–262. https://doi.org/10.1093/icesjms/fsab261
Andrews, A. J., Pampoulie, C., Di Natale, A., Addis, P., Bernal-Casasola, D., Aniceti, V., Carenti, G., Gómez-Fernández, V., Chosson, V., Ughi, A., Von Tersch, M., Fontanals-Coll, M., Cilli, E., Onar, V., Tinti, F., & Alexander, M. (2023). Exploitation shifted trophic ecology and habitat preferences of Mediterranean and Black Sea bluefin tuna over centuries. Fish and Fisheries, 24(6), 1067–1083. https://doi.org/10.1111/faf.12785
Andrews, A. J., Puncher, G. N., Bernal-Casasola, D., Di Natale, A., Massari, F., Onar, V., Toker, N. Y., Hanke, A., Pavey, S. A., Savojardo, C., Martelli, P. L., Casadio, R., Cilli, E., Morales-Muñiz, A., Mantovani, B., Tinti, F., & Cariani, A. (2021). Ancient DNA SNP-panel data suggests stability in bluefin tuna genetic diversity despite centuries of fluctuating catches in the eastern Atlantic and Mediterranean. Scientific Reports, 11(1), 20744. https://doi.org/10.1038/s41598-021-99708-9
Anonymous. (1875). The hand book to Monterey and vicinity: Containing a brief resumé of the history of Monterey since its discovery; a general review of the resources and products of Monterey and the county; descriptive sketches of the town, and the points of interest in the neighborhood; Carmel Mission and Valley; Pacific Grove Retreat; Point Cypress, Point Pinos and the light house, Salinas, Castroville, San Juan, San Antonio Mission, and other places of interest in the county, a complete guide book for tourists, campers and visitors. Walton & Curtis.
Armentrout-Ma, L. E. (1981). Chinese in California’s fishing industry, 1850–1941. California History, 60(2), 142–157. https://doi.org/10.2307/25158037
Badenhorst, S., & Ross, D. (2019). The fauna from Lion Island, a late nineteenth and early twentieth century Chinese community in British Columbia, Canada. In Henan Provincial Institute of Cultural Heritage and Archaeology (Ed.), Zooarchaeology, volume 3: Proceedings of Zhengzhou 2016 International Committee Meeting of ICAZ and “Archaeozoology: Global Developments and Chinese Perspectives” (pp. 46–58). Cultural Relics Press.
Barrett, J. H. (2019). An environmental (pre)history of European fishing: Past and future archaeological contributions to sustainable fisheries. Journal of Fish Biology, 94(6), 1033–1044. https://doi.org/10.1111/jfb.13929
Barrett, J. H., Locker, A. M., & Roberts, C. M. (2004). 'Dark Age economics' revisited: The English fish-bone evidence, AD 600–1600. Antiquity, 78(301), 618–636. https://doi.org/10.1017/S0003598X00113262
Bas, M., Salemme, M., Green, E. J., Santiago, F., Speller, C., Álvarez, M., Briz i Godino, I., & Cardona, L. (2020). Predicting habitat use by the Argentine hake Merluccius hubbsi in a warmer world: Inferences from the Middle Holocene. Oecologia, 193(2), 461–474. https://doi.org/10.1007/s00442-020-04667-z
Bentz, L., & Braje, T. J. (2020). Bounty from the sea: Chinese foundations of the commercial shrimp, squid, and abalone fisheries in California. In C. Rose & J. R. Kennedy (Eds.), Chinese diaspora archaeology in North America (pp. 275–305). University Press of Florida. https://doi.org/10.5744/florida/9780813066356.003.0012
Braje, T. J., & Bentz, L. (2015). The Archaeology of overseas Chinese abalone fishermen on Santa Rosa Island, Alta California. Journal of Chinese Overseas, 11(1), 87–103. https://doi.org/10.1163/17932548-12341299
Braje, T. J., Rick, T. C., Szpak, P., Newsome, S. D., McCain, J. M., Elliott Smith, E. A., Glassow, M., & Hamilton, S. L. (2017). Historical ecology and the conservation of large, hermaphroditic fishes in Pacific Coast kelp forest ecosystems. Science Advances, 3(2), e1601759. https://doi.org/10.1126/sciadv.1601759
Burton, E. J., & Lea, R. N. (2019). Annotated checklist of fishes from Monterey Bay National Marine Sanctuary with notes on extralimital species. ZooKeys, 887, 1–119. https://doi.org/10.3897/zookeys.887.38024
Byers, R. D. (1940). The California shark fishery. California Fish and Game, 26(1), 23–38.
Carlisle, A. B., & Starr, R. M. (2009). Habitat use, residency, and seasonal distribution of female leopard sharks Triakis semifasciata in Elkhorn Slough, California. Marine Ecology Progress Series, 380, 213–228. https://doi.org/10.3354/meps07907
Chee-Beng, T. (2011). Introduction. In T. Chee-Beng (Ed.), Chinese food and foodways in Southeast Asia and beyond (pp. 1–19). NUS Press.
Chiang, C. Y. (2004). Monterey-by-the-Smell: Odors and social conflict on the California coastline. Pacific Historical Review, 73(2), 183–214. https://doi.org/10.1525/phr.2004.73.2.183
Collins, J. W. (1892). Report on the Pacific Coast of the United States. In M. McDonald (Ed.), Report of the Commissioner of Fish and Fisheries for 1888 [July 1, 1888, to June 30, 1889.], part 16 (pp. 3–269). Government Printing Office.
Cooper, A., & Poinar, H. N. (2000). Ancient DNA: Do it right or not at all. Science, 289(5482), 1139. https://doi.org/10.1126/science.289.5482.1139b
Cummings, L. S., Voss, B. L., Yu, C. Y., Kováčik, P., Puseman, K., Yost, C., Kennedy, R., & Kane, M. S. (2014). Fan and tsai: Intracommunity variation in plant-based food consumption at the Market Street Chinatown, San Jose, California. Historical Archaeology, 48(2), 143–172. https://doi.org/10.1007/BF03376931
Dabney, J., Meyer, M., & Pääbo, S. (2013). Ancient DNA damage. Cold Spring Harbor Perspectives in Biology, 5(7), a012567. https://doi.org/10.1101/cshperspect.a012567
Daily Alta California. (1866). Oriental banquet in honor of the American ministers to China and Japan. Daily Alta California, 18(5927), June 2. San Francisco.
Daily Alta California. (1887). The fish commission. Daily Alta California, 42(13798), June 11, 1. San Francisco.
Daily Humboldt Times. (1887). Mrs. Shepherd’s Chinese lunch. Daily Humboldt Times, 28(132), December 3. Eureka.
Daily Los Angeles Herald. (1900). A Chinese dinner. Daily Los Angeles Herald, 27(315), August 12, 26. Los Angeles.
Douglass, K., Antonites, A. R., Quintana Morales, E. M., Grealy, A., Bunce, M., Bruwer, C., & Gough, C. (2018). Multi-analytical approach to zooarchaeological assemblages elucidates Late Holocene coastal lifeways in southwest Madagascar. Quaternary International, 471(Part A), 111–131. https://doi.org/10.1016/j.quaint.2017.09.019
Ebert, D. A. (2003). Sharks, rays, and chimaeras of California. University of California Press.
Elliott Smith, E. A., Szpak, P., Braje, T. J., Newsom, B., & Rick, T. C. (2023). Pre-industrial ecology and foraging behavior of Swordfish Xiphias gladius in the eastern North Pacific. Marine Ecology Progress Series, 711, 129–134. https://doi.org/10.3354/meps14305
Emery, M. V., Bolhofner, K., Ghafoor, S., Winningear, S., Buikstra, J. E., Fulginiti, L. C., & Stone, A. C. (2022). Whole mitochondrial genomes assembled from thermally altered forensic bones and teeth. Forensic Science International: Genetics, 56, 102610. https://doi.org/10.1016/j.fsigen.2021.102610
Evening Sentinel. (1907). Friends of Woo Gap are invited to marvelous New Year feast. Evening Sentinel, 11(221), February 20, 6. Santa Cruz.
Fields, A. T., Abercrombie, D. L., Eng, R., Feldheim, K., & Chapman, D. D. (2015). A novel mini-DNA barcoding assay to identify processed fins from internationally protected shark species. PLoS ONE, 10(2), e0114844. https://doi.org/10.1371/journal.pone.0114844
Folkard, H. C. (1901). The sailing boat: A treatise on sailing boats and small yachts, their varieties of type, sails, rig, &c. with practical instructions in sailing and management, also the one-design and restricted classes, fishing and shooting boats, sailing chariots and ice yachts, foreign and colonial boats, canoes, &c., &c (5th ed.). Edward Stanford.
Folmer, O., Black, M., Hoeh, W., Lutz, R., & Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3(5), 249–299. https://doi.org/10.1071/ZO9660275
Fong, K. N., Ng, L. W., Lee, J., Peterson, V. L., & Voss, B. L. (2022). Race and racism in archaeologies of Chinese American communities. Annual Review of Anthropology, 51, 233–250. https://doi.org/10.1146/annurev-anthro-041320-014548
Free, C. M., Vargas Poulsen, C., Bellquist, L. F., Wassermann, S. N., & Oken, K. L. (2022). The CALFISH database: A century of California’s non-confidential fisheries landings and participation data. Ecological Informatics, 69, 101599. https://doi.org/10.1016/j.ecoinf.2022.101599
Freeman, M. (1977). Sung. In K. C. Chang (Ed.), Food in Chinese culture: Anthropological and historical perspectives (pp. 141–192). Yale University Press.
Gobalet, K. W. (2001). A critique of faunal analysis; Inconsistency among experts in blind tests. Journal of Archaeological Science, 28(4), 377–386. https://doi.org/10.1006/jasc.2000.0564
Gobalet, K. W., Schulz, P. D., Wake, T. A., & Siefkin, N. (2004). Archaeological perspectives on Native American fisheries of California, with emphasis on steelhead and salmon. Transactions of the American Fisheries Society, 133(4), 801–833. https://doi.org/10.1577/T02-084.1
Grealy, A., Douglass, K., Haile, J., Bruwer, C., Gough, C., & Bunce, M. (2016). Tropical ancient DNA from bulk archaeological fish bone reveals the subsistence practices of a historic coastal community in southwest Madagascar. Journal of Archaeological Science, 75, 82–88. https://doi.org/10.1016/j.jas.2016.10.001
Guiry, E. J., Kennedy, J. R., O’Connell, M. T., Gray, D. R., Grant, C., & Szpak, P. (2021). Early evidence for historical overfishing in the Gulf of Mexico. Science Advances, 7(32), eabh2525. https://doi.org/10.1126/sciadv.abh2525
Guiry, E. J., Buckley, M., Orchard, T. J., Hawkins, A. L., Needs-Howarth, S., Holm, E., & Szpak, P. (2020a). Deforestation caused abrupt shift in Great Lakes nitrogen cycle. Limnology and Oceanography, 65(8), 1921–1935. https://doi.org/10.1002/lno.11428
Guiry, E., Royle, T. C. A., Matson, R. G., Ward, H., Weir, T., Waber, N., Brown, T. J., Hunt, B. P. V., Price, M. H. H., Finney, B. P., Kaeriyama, M., Qin, Y., Yang, D. Y., & Szpak, P. (2020c). Differentiating salmonid migratory ecotypes through stable isotope analysis of collagen: Archaeological and ecological applications. PLoS ONE, 15(4), e0232180. https://doi.org/10.1371/journal.pone.0232180
Guiry, E. J., Royle, T. C. A., Orchard, T. J., Needs-Howarth, S., Yang, D. Y., & Szpak, P. (2020b). Evidence for freshwater residency among Lake Ontario Atlantic salmon (Salmo salar) spawning in New York. Journal of Great Lakes Research, 46(4), 1036–1043. https://doi.org/10.1016/j.jglr.2020.05.009
Hawkins, A. L., Needs-Howarth, S., Orchard, T. J., & Guiry, E. J. (2019). Beyond the local fishing hole: A preliminary study of pan-regional fishing In southern Ontario (ca. 1000 CE to 1750 CE). Journal of Archaeological Science: Reports, 24, 856–868. https://doi.org/10.1016/j.jasrep.2019.03.007
Hebert, P. D. N., Cywinska, A., Ball, S. L., & deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences, 270(1512), 313–321. https://doi.org/10.1098/rspb.2002.2218
Hsu, M. (2006). Trading with Gold Mountain: Jinshanzhuang and networks of kinship and native place. In S. Chan (Ed.), Chinese American transnationalism: The flow of people, resources, and ideas between China and America during the Exclusion Era (pp. 22–33). Temple University Press.
Jackson, J. B. C., Kirby, M. X., Berger, W. H., Bjorndal, K. A., Botsford, L. W., Bourque, B. J., Bradbury, R. H., Cooke, R., Erlandson, J., Estes, J. A., Hughes, T. P., Kidwell, S., Lange, C. B., Lenihan, H. S., Pandolfi, J. M., Peterson, C. H., Steneck, R. S., Tegner, M. J., & Warner, R. R. (2001). Historical overfishing and the recent collapse of coastal ecosystems. Science, 293(5530), 629–638. https://doi.org/10.1126/science.1059199
Johnson, B. M., Kemp, B. M., & Thorgaard, G. H. (2018). Increased mitochondrial DNA diversity in ancient Columbia River basin Chinook salmon Oncorhynchus tshawytscha. PLoS ONE, 13(1), e0190059. https://doi.org/10.1371/journal.pone.0190059
Jordan, D. S. (1887a), The Chinese fishermen of the Pacific Coast. In G. Brown Good (Ed.), The fisheries and fishery industries of the United States, section III: The fishing grounds of North America with forty-nine charts (pp. 37–42). Government Printing Office.
Jordan, D. S. (1887b). The fisheries of the Pacific Coast. In G. Brown Good (Ed.), The fisheries and fishery industries of the United States, section III: The fishing grounds of North America with forty-nine charts (pp. 591–630). Government Printing Office.
Jordan, D. S. (1905). A guide to the study of fishes, volume 1. Henry Holt and Company.
Jordan, D. S. (1892). The Hopkins Seaside Laboratory. Science, 20(496), 76–77. https://doi.org/10.1126/science.ns-20.496.76
Jordan, D. S., & Gilbert, C. H. (1882). Notes on the fishes of the Pacific Coast of the United States. Proceedings of the United States National Museum, 4(191), 29–70.
Kennedy, J. R., & Rose, C. (2020). Charting a new course for Chinese diaspora archaeology in North America. In C. Rose & J. R. Kennedy (Eds.), Chinese diaspora archaeology in North America (pp. 1–34). University Press of Florida. https://doi.org/10.5744/florida/9780813066356.003.0001
Kennedy, J. R. (2015). Zooarchaeology, localization, and Chinese railroad workers in North America. Historical Archaeology, 49(1), 122–133. https://doi.org/10.1007/BF03376963
Kennedy, J. R. (2017). The fresh and the salted: Chinese migrant fisheries engagement and trade in nineteenth-century North America. Journal of Ethnobiology, 37(3), 421–439. https://doi.org/10.2993/0278-0771-37.3.421
Kennedy, J. R., Bingham, B., Flores, M. F., & Kemp, B. M. (2022). Ancient DNA identification of giant snakehead (Channa micropeltes) remains from the Market Street Chinatown and some implications for the nineteenth-century Pacific World fish trade. American Antiquity, 87(1), 42–58. https://doi.org/10.1017/aaq.2021.61
Kennedy, J. R., & Guiry, E. J. (2023). Exploring railroad impacts on meat trade: An isotopic investigation of meat sourcing and animal husbandry at Chinese diaspora sites in the American West. International Journal of Historical Archaeology, 27(2), 393–423. https://doi.org/10.1007/s10761-022-00663-6
Kennedy, J. R., Rogers, L., & Kaestle, F. A. (2018). Ancient DNA evidence for the regional trade of bear paws by Chinese diaspora communities in 19th-century western North America. Journal of Archaeological Science, 99, 135–142. https://doi.org/10.1016/j.jas.2018.09.005
Kohrs, D. (2013). Hopkins Seaside Laboratory (1892 -1917): A summer school of science. Stanford University Libraries. Retrieved September 23, 2023, from https://exhibits.stanford.edu/seaside
Lambrides, A. B. J., McNiven, I. J., & Ulm, S. (2019). Meta-analysis of Queensland’s coastal Indigenous fisheries: Examining the archaeological evidence for geographic and temporal patterning. Journal of Archaeological Science: Reports, 28, 102057. https://doi.org/10.1016/j.jasrep.2019.102057
Lee, L. A. (2006). History Rewritten: The story of Quock Mui Jeung. Asian Pacific American Law Journal, 11(1), 75–91.
Leonard, J. A., Shanks, O., Hofreiter, M., Kreuz, E., Hodges, L., Ream, W., Wayne, R. K., & Fleischer, R. C. (2007). Animal DNA in PCR reagents plagues ancient DNA research. Journal of Archaeological Science, 34(9), 1361–1366. https://doi.org/10.1016/j.jas.2006.10.023
Los Angeles Herald. (1904). Pong Wong Wuey has 1200 members. Los Angeles Herald, 31(97), August 12, 8. Los Angeles.
Love, M. S., Bizzarro, J. S., Cornthwaite, A. M., Frable, B. W., & Maslenikov, K. P. (2021). Checklist of marine and estuarine fishes from the Alaska–Yukon border, Beaufort Sea, to Cabo San Lucas, Mexico. Zootaxa, 5053(1), 1–285. https://doi.org/10.11646/zootaxa.5053.1.1
Lydon, S. (1985). Chinese gold: The Chinese in the Monterey Bay region. Capitola Book Company.
Lydon, S. (1997). The Japanese in the Monterey Bay region: A brief history. Capitola Book Company.
Martínez-García, L., Ferrari, G., Oosting, T., Ballantyne, R., van der Jagt, I., Ystgaard, I., Harland, J., Nicholson, R., Hamilton-Dyer, S., Baalsrud, H. T., Brieuc, M. S. O., Atmore, L. M., Burns, F., Schmölcke, U., Jakobsen, K. S., Jentoft, S., Orton, D., Hufthammer, A. K., Barrett, J. H., & Star, B. (2021). Historical demographic processes dominate genetic variation in ancient Atlantic Cod mitogenomes. Frontiers in Ecology and Evolution, 9, 671281. https://doi.org/10.3389/fevo.2021.671281
McKechnie, I., & Moss, M. L. (2016). Meta-analysis in zooarchaeology expands perspectives on Indigenous fisheries of the Northwest Coast of North America. Journal of Archaeological Science: Reports, 8, 470–485. https://doi.org/10.1016/j.jasrep.2016.04.006
Miszaniec, J. I., Ramirez, M., Morales, J., Canzonieri, C., & Eerkens, J. W. (2021). Use of archaeological data in retracing diet and growth of extirpated fish populations in the California Delta: An allometric and isotopic approach to Sacramento perch (Archoplites interruptus) historical ecology. Journal of Archaeological Science: Reports, 39, 103191. https://doi.org/10.1016/j.jasrep.2021.103191
Monks, G. M. (1987). Prey as bait: The Deep Bay example. Canadian Journal of Archaeology, 11, 119–142.
Monterey Gazette. (1864). John Chinadom. Monterey Gazette, 1(6), January 15. Monterey.
Palmer, E., Tushingham, S., & Kemp, B. M. (2018). Human use of small forage fish: Improved ancient DNA species identification techniques reveal long term record of sustainable mass harvesting of smelt fishery in the northeast Pacific Rim. Journal of Archaeological Science, 99, 143–152. https://doi.org/10.1016/j.jas.2018.09.014
Pauly, D. (1995). Anecdotes and the shifting baseline syndrome of fisheries. Trends in Ecology & Evolution, 10(10), 430. https://doi.org/10.1016/S0169-5347(00)89171-5
Popper, V. S. (2020). Flexible plant food practices among the nineteenth-century Chinese migrants to western North America. In C. Rose & J.R. Kennedy (Eds.), Chinese diaspora archaeology in North America (pp. 306–333). University Press of Florida. https://doi.org/10.5744/florida/9780813066356.003.0013
Porcasi, J. F. (2017). Persistence of traditional lifeways by early Chinese immigrants: Faunal evidence from the High Lung Laundry (CA-SBA-2752H) Santa Barbara, California. International Journal of Historical Archaeology, 21(3), 558–576. https://doi.org/10.1007/s10761-016-0383-0
Ratnasingham, S., & Hebert, P. D. N. (2007). BOLD: Barcode of Life Data System. Molecular Ecology Notes, 7(3), 355–364. https://doi.org/10.1111/j.1471-8286.2006.01678.x
Rick, T. C., Erlandson, J. M., Glassow, M. A., & Moss, M. L. (2002). Evaluating the economic significance of sharks, skates, and rays (elasmobranchs) in prehistoric economies. Journal of Archaeological Science, 29(2), 111–122. https://doi.org/10.1006/jasc.2000.0637
Ripley, W. S. (1946). The soupfin shark and the fishery. California Division of Fish and Game Fish Bulletin, 64, 7–37.
Sayers, E. W., Cavanaugh, M., Clark, K., Pruitt, K. D., Sherry, S. T., Yankie, L., & Karsch-Mizrachi, I. (2023). GenBank 2023 update. Nucleic Acids Research, 51(D1), D141–D144. https://doi.org/10.1093/nar/gkac1012
Schulz, P. D. (2002). Fish remains from the Woolen Mills Chinatown site, San Jose, California. In R. Allen, R. S. Baxter, A. Medin, J. G. Costello, & C. Y. Young (Eds.), Excavation of the Woolen Mills Chinatown (CA-SCL-807H), San Jose, volume 2. Report submitted to the California Department of Transportation, Oakland.
Seersholm, F. V., Cole, T. L., Grealy, A., Rawlence, N. J., Greig, K., Knapp, M., Stat, M., Hansen, A. J., Easton, L. J., Shepherd, L., Tennyson, A. J. D., Scofield, R. P., Walter, R., & Bunce, M. (2018). Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey. Proceedings of the National Academy of Sciences of the United States of America, 115(30), 7771–7776. https://doi.org/10.1073/pnas.1803573115
Shepherd, L. D., & Campbell, M. (2021). Ancient DNA analysis of an archaeological assemblage of Chondrichthyes vertebrae from South Auckland, New Zealand. Journal of Archaeological Science: Reports, 36,
Simoons, F. J. (1991). Food in China. CRC Press.
Smith, S. E., & Susumu, K. (1979). The fisheries of San Francisco Bay: Past, present and future. In T. J. Conomos (Ed.), San Francisco Bay: The urbanized estuary: Investigations into the natural history of San Francisco Bay and Delta with reference to the influence of man (pp. 445–468). Pacific Division of the American Association for the Advancement of Science c/o California Academy of Sciences.
Speller, C. F., Hauser, L., Lepofsky, D., Moore, J., Rodrigues, A. T., Moss, M. L., McKechnie, I., & Yang, D. Y. (2012). High potential for using DNA from ancient herring bones to inform modern fisheries management and conservation. PLoS ONE, 7(11), e51122. https://doi.org/10.1371/journal.pone.0051122
Spence, J. (1977). Ch’ing. In K. C. Chang (Ed.), Food in Chinese culture: Anthropological and historical perspectives (pp. 259–294). Yale University Press.
Sunseri, C. K. (2020). Meat economies of the Chinese American West. In C. Rose & J. R. Kennedy (Eds.), Chinese diaspora archaeology in North America (pp. 250–274). University Press of Florida. https://doi.org/10.5744/florida/9780813066356.003.0011
Van Tilburg, H. K. (2007). Vessels of exchange: The global shipwright in the Pacific. In J. H. Bentley, R. Bridenthal R, & K. Wigen (Eds.), Seascapes: Maritime histories, littoral cultures, and transoceanic exchanges (pp. 38–52). University of Hawaii Press.
Vannuccini, S. (1999). Shark utilization, marketing and trade. Food and Agriculture Organization of the United Nations.
Voss, B. L., Kennedy, J. R., Tan, J., (S.), & Ng, L. (2018). The archaeology of home: Qiaoxiang and nonstate actors in the archaeology of the Chinese diaspora. American Antiquity, 83(3), 407–426. https://doi.org/10.1017/aaq.2018.16
Walford, L. A. (1931). The sharks and rays of California. California Division of Fish and Game Fish Bulletin, 45, 3–66.
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R., & Hebert, P. D. N. (2005). DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society B: Biological Sciences, 360(1462), 1847–1857. https://doi.org/10.1098/rstb.2005.1716
Wave, S. F. (1897). The Monterey fisheries. Evening Sentinel, 1(227), February 20. Santa Cruz.
West, G. J., & Stevens, J. D. (2001). Archival tagging of school shark, Galeorhinus galeus, in Australia: Initial results. Environmental Biology of Fishes, 60(1–3), 283–298. https://doi.org/10.1023/A:1007697816642
Williams, B., (2011). The archaeology of objects and identities at the Point Alones Chinese village, Pacific Grove, CA (1860– 1906). Ph.D. dissertation. Stanford University.
Winn, J. C., Maduna, S. N., & Bester-van der Merwe, A. E. (2024). A comprehensive phylogenomic study unveils evolutionary patterns and challenges in the mitochondrial genomes of Carcharhiniformes: A focus on Triakidae. Genomics, 116(1)
Winters, M., Barta, J. L., Monroe, C., & Kemp, B. M. (2011). To clone or not to clone: method analysis for retrieving consensus sequences in ancient DNA samples. PLoS ONE, 6(6)
Yang, D. Y., Cannon, A., & Saunders, S. R. (2004). DNA species identification of archaeological salmon bone from the Pacific Northwest Coast of North America. Journal of Archaeological Science, 31(5), 619–631. https://doi.org/10.1016/j.jas.2003.10.008
Yang, D. Y., Eng, B., Waye, J. S., Dudar, J. C., & Saunders, S. R. (1998). Improved DNA extraction from ancient bones using silica-based spin columns. American Journal of Physical Anthropology, 105(4), 539–543. https://doi.org/10.1002/(SICI)1096-8644(199804)105:4%3c539::AID-AJPA10%3e3.0.CO;2-1
Yang, D. Y., Liu, L., Chen, X., & Speller, C. F. (2008). Wild or domesticated: DNA analysis of ancient water buffalo remains from north China. Journal of Archaeological Science, 35(10), 2778–2785. https://doi.org/10.1016/j.jas.2008.05.010
Yang, D. Y., & Watt, K. (2005). Contamination controls when preparing archaeological remains for ancient DNA analysis. Journal of Archaeological Science, 32(3), 331–336. https://doi.org/10.1016/j.jas.2004.09.008
Yoklavich, M. M., Cailliet, G. M., Barry, J. P., Ambrose, D. A., & Antrim, B. S. (1991). Temporal and spatial patterns in abundance and diversity of fish assemblages in Elkhorn Slough, California. Estuaries, 14(4), 465–480. https://doi.org/10.2307/1352270
Yu, H. (2020). Multisited networks: The underlying analytical power of transnational and diasporic approaches. In C. Rose & J. R. Kennedy (Eds.), Chinese diaspora archaeology in North America (pp. 334–350). University Press of Florida. https://doi.org/10.5744/florida/9780813066356.003.0014
Acknowledgements
Our thanks go to Dr. Laura Jones, Stanford University Heritage Services, and Stanford University Archaeology Collections for providing access to the Point Alones faunal assemblage and permissions to conduct destructive analyses. We are also grateful to Dr. Barbara Voss (Department of Anthropology, Stanford University) for her continued support of research on Chinese diaspora collections. We also wish to recognize the late Gerry Low-Sabado, a descendant of Point Alones’ inhabitants, for their tirelessly promotion of Chinese heritage in Monterey Bay and generously sharing her family’s history with us. We thank Nate King and the Pacific Grove Museum of Natural History (https://www.pgmuseum.org) as well as the Bancroft Library at the University of California, Berkeley (https://www.lib.berkeley.edu/visit/bancroft) for providing copies of archival photographs of Point Alones held in their collections. We also acknowledge the use of a digital reproduction of a historical image produced by the University of Washington Libraries Freshwater Marine Image Bank (https://content.lib.washington.edu/fishweb/index.html). Interpreting our data would also not have been possible without the use of the digitized historical newspapers in the California Digital Newspaper Collection hosted by Center for Bibliographical Studies and Research at University of California, Berkley (https://cdnc.ucr.edu/). We thank Dr. Hua Zhang for technical assistance and her diligent management of the Simon Fraser University Ancient DNA Laboratory. We also thank members of the International Council of Archaeozoology Fish Remains Working Group and Simon Fraser University Department of Archaeology and School of Environmental Science for comments on earlier presentations of this study. Royle would also like to express his thanks to his postdoctoral supervisor —Dr. Jonathan Driver (Department of Archaeology, Simon Fraser University)—for his feedback on this manuscript and encouraging him to pursue this side project during his postdoctoral fellowship.
Funding
Open access funding provided by the NTNU Norwegian University of Science and Technology. This study was supported by a Small Grant from the Save Our Seas Foundation (Project number: SOSF 569) and funding from Stanford University’s Lang Fund for Environmental Anthropology.
Author information
Authors and Affiliations
Contributions
Thomas C.A. Royle: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Data curation, Visualization, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition, J. Ryan Kennedy: Conceptualization, Resources, Writing - review & editing, Project administration, Funding acquisition, Eric J. Guiry: Conceptualization, Writing - review & editing, Project administration, Funding acquisition, Luke S. Jackman: Investigation, Validation, Writing - review & editing, Yuka Shichiza: Investigation, Validation, Writing - review & editing, Dongya Y. Yang: Resources, Supervision, Writing - review & editing.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Royle, T.C.A., Kennedy, J.R., Guiry, E.J. et al. Sharkaeology: Expanding Understandings of Historical Chinese Diaspora Shark Fisheries in Monterey Bay, California, through the Genetic Species Identification of Archaeological Chondrichthyes Remains. Hum Ecol (2024). https://doi.org/10.1007/s10745-024-00521-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s10745-024-00521-5