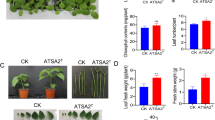
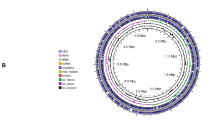
Abstract
Leclercia adecarboxylata QDSM01 is a plant growth-promoting bacterium isolated from the rhizosphere soil of maize. However, the relevant molecular mechanisms remain obscure. This study investigated the effect of Leclercia adecarboxylata QDSM01 on the seedling growth of maize, rice and soybean and uncovered the bacterium’s biochemical characterization (biofilm, plant hormone secretion, nitrogen generation, phosphorus and potassium solubilization). In-depth analysis of the functional genes related to the biochemical characterization was conducted by complete genome sequence. Strain QDSM01 significantly promoted the seedling growth of maize and rice, but not soybean. The strain QDSM01 was identified as Leclercia adecarboxylata based on a comparison of 16 S rRNA gene sequences and complete genome sequence. The complete genome sequence indicated the strain comprised a 4,461,951 bp chromosome and 5 plasmids. Moreover, antiSMASH analysis revealed a total of four secondary metabolite gene clusters, consisting of putative non-ribosomal peptide synthase (NRPS), terpenoids, thiopeptides and arylpolyene. These gene clusters play an important role in promoting plant growth and resistance. Furthermore, the genes involved in biofilm formation, quorum sensing, chemotaxis, motility, indole-3-acetic acid production (IAA), siderophore, nitrogen generation, solubilization and uptake of phosphate and potassium were identified. Meanwhile, in vitro experiments were also performed to confirm these functions. In addition, the strong IAA production ability of strain QDSM01 was observed than other strains of Leclercia adecarboxylata. These results suggest that Leclercia adecarboxylata QDSM01 can serve as a biofertilizer that improves plant growth. This study will be helpful for further studies of Leclercia adecarboxylata on the mechanisms of plant growth promotion.
Graphical abstract







Similar content being viewed by others
Data availability
The genome sequences and associated data for Leclercia adecarboxylata QDSM01 reported in this study were deposited in NCBI under the BioProject accession number PRJNA841055.
References
Abdelaal K, AlKahtani M, Attia K, Hafez Y, Király L, Künstler A (2021) The role of plant growth-promoting bacteria in alleviating the adverse effects of drought on plants. Biology 10:520. https://doi.org/10.3390/biology10060520
Andrés-Barrao C, Lafi FF, Alam I, de Zélicourt A, Eida AA, Bokhari A et al (2017) Complete genome sequence analysis of enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front Microbiol 8:2023. https://doi.org/10.3389/fmicb.2017.02023
Berendsen RL, Vismans G, Yu K, Song Y, de Jonge R, Burgman WP et al (2018) Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J 12:1496–1507. https://doi.org/10.1038/s41396-018-0093-1
Bhise KK, Dandge PB (2019) Mitigation of salinity stress in plants using plant growth promoting bacteria. Symbiosis 79:191–204. https://doi.org/10.1007/s13199-019-00638-y
Bouteiller M, Dupont C, Bourigault Y, Latour X, Barbey C, Konto-Ghiorghi Y et al (2021) Pseudomonas flagella: generalities and specificities. Int J Mol Sci 22:3337. https://doi.org/10.3390/ijms22073337
Chen YF, Yang H, Shen ZZ, Ye JR (2022) Whole-genome sequencing and potassium-solubilizing mechanism of Bacillus aryabhattai SK1-7. Front Microbiol 12:722379. https://doi.org/10.3389/fmicb.2021.722379
Crits-Christoph A, Bhattacharya N, Olm MR, Song YS, Banfield JF (2020) Transporter genes in biosynthetic gene clusters predict metabolite characteristics and siderophore activity. Genome Res 31:239–250. https://doi.org/10.1101/gr.268169.120
Cueva-Yesquén LG, Goulart MC, de Angelis A, Nopper Alves D, Fantinatti-Garboggini M (2021) Multiple plant growth-promotion traits in endophytic bacteria retrieved in the vegetative stage from passionflower. Front Plant Sci 11:2282. https://doi.org/10.3389/fpls.2020.621740
Dahmani MA, Desrut A, Moumen B, Verdon J, Mermouri L, Kacem M et al (2020) Unearthing the plant growth-promoting traits of Bacillus megaterium RmBm31, an endophytic bacterium isolated from root nodules of Retama monosperma. Front Plant Sci 11:124. https://doi.org/10.3389/fpls.2020.00124
Danish S, Kiran S, Fahad S, Ahmad N, Ali MA, Tahir FA et al (2019) Alleviation of chromium toxicity in maize by Fe fortification and chromium tolerant ACC deaminase producing plant growth promoting rhizobacteria. Ecotoxicol Environ Saf 185:109706. https://doi.org/10.1016/j.ecoenv.2019.109706
Danish S, Zafar-ul-Hye DM, Mohsin F, Hussain M (2020) ACC-deaminase producing plant growth promoting rhizobacteria and biochar mitigate adverse effects of drought stress on maize growth. PLoS ONE 15:e0230615. https://doi.org/10.1371/journal.pone.0230615
de Souza RSC, Armanhi JSL, Damasceno NdB, Imperial J, Arruda P (2019) Genome sequences of a plant beneficial synthetic bacterial community reveal genetic features for successful plant colonization. Front Microbiol 10:1779. https://doi.org/10.3389/fmicb.2019.01779
Dong X, Tu C, Xie Z, Luo Y, Zhang L, Li Z (2022) The genome of Bacillus velezensis SC60 provides evidence for its plant probiotic effects. Microorganisms 10:767. https://doi.org/10.3390/microorganisms10040767
Etesami H, Emami S, Alikhani HA (2017) Potassium solubilizing bacteria (KSB): mechanisms, promotion of plant growth, and future prospects—a review. J Soil Sci Plant Nutr 17:897–911. https://doi.org/10.4067/S0718-95162017000400005
Fong JCN, Syed KA, Klose KE, Yildiz FH (2010) Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiol 156:2757–2769. https://doi.org/10.1099/mic.0.040196-0
Gu S, Wei Z, Shao Z, Friman VP, Cao K, Yang T et al (2020) Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat Microbiol 5:1002–1010. https://doi.org/10.1038/s41564-020-0719-8
Guan J, Xiao X, Xu S, Gao F, Wang J, Wang T et al (2015) Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J Microbiol 53:633–642. https://doi.org/10.1007/s12275-015-0099-6
Iqbal S, Ullah N, Janjua HA (2021) In vitro evaluation and genome mining of Bacillus subtilis strain RS10 reveals its biocontrol and plant growth-promoting potential. Agriculture 11:1273. https://doi.org/10.3390/agriculture11121273
Jung BK, Khan AR, Hong S-J, Park G-S, Park Y-J, Kim H-J et al (2017) Quorum sensing activity of the plant growth-promoting rhizobacterium Serratia glossinae GS2 isolated from the sesame (Sesamum indicum L.) rhizosphere. Ann Microbiol 67:623–632. https://doi.org/10.1007/s13213-017-1291-1
Kang S, Asaf S, Kim S-J, Yun B-W, Lee I-J (2016) Complete genome sequence of plant growth-promoting bacterium Leifsonia xyli SE134, a possible gibberellin and auxin producer. J Biotechnol 239:34–38. https://doi.org/10.1016/j.jbiotec.2016.10.004
Kang SM, Shahzad R, Bilal S, Khan AL, Park YG, Lee KE et al (2019) Indole-3-acetic-acid and ACC deaminase producing Leclercia adecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol 19:80. https://doi.org/10.1186/s12866-019-1450-6
Kang S, Shahzad R, Khan MA, Hasnain Z, Lee K-E, Park H-S et al (2021) Ameliorative effect of indole-3-acetic acid and siderophore-producing Leclercia adecarboxylata MO1 on cucumber plants under zinc stress. J Plant Interact 16:30–41. https://doi.org/10.1080/17429145.2020.1864039
Kumawat KC, Sharma P, Singh I, Sirari A, Gill BS (2019) Co-existence of Leclercia adecarboxylata (LSE-1) and Bradyrhizobium sp. (LSBR-3) in nodule niche for multifaceted effects and profitability in soybean production. World J Microbiol Biotechnol 35:172. https://doi.org/10.1007/s11274-019-2752-4
López D, Fischbach MA, Chu F, Losick R, Kolter R (2009) Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci 106:280–285
Lulamba TE, Green E, Serepa-Dlamini MH (2021) Genome assembly and annotation of Photorhabdus heterorhabditis strain ETL reveals genetic features involved in pathogenicity with its associated entomopathogenic nematode and anti-host effectors with biocontrol potential applications. Gene 795:145780. https://doi.org/10.1016/j.gene.2021.145780
Naveed M, Ahmed I, Khalid N, Mumtaz AS (2014) Bioinformatics based structural characterization of glucose dehydrogenase (gdh) gene and growth promoting activity of Leclercia sp. QAU-66. Brazilian J Microbiol 45:603–611. https://doi.org/10.1590/s1517-83822014000200031
Niu B, Paulson Joseph N, Zheng X, Kolter R (2017) Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci 114:E2450–E2459
Qiao Y, Feng L, Jia R, Luo Y, Yang Q (2022) Motility, biofilm formation and associated gene expression in Vibrio parahaemolyticus impaired by co-culture with live Ulva fasciata. J Appl Microbiol 132:101–112. https://doi.org/10.1111/jam.15175
Reitz ZL, Sandy M, Butler A (2017) Biosynthetic considerations of triscatechol siderophores framed on serine and threonine macrolactone scaffolds. Metallomics 9:824–839. https://doi.org/10.1039/c7mt00111h
Romero D, de Vicente A, Rakotoaly RH, Dufour SE, Veening J-W, Arrebola E et al (2007) The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol Plant Microbe Interact 20:430–440. https://doi.org/10.1094/MPMI-20-4-0430
Sandilya SP, Jeevan B, Subrahmanyam G, Dutta K, Vijay N, Bhattacharyya N et al (2022) Co-inoculation of native multi-trait plant growth promoting rhizobacteria promotes plant growth and suppresses alternaria blight disease in castor (Ricinus communis L). Heliyon 8:e11886. https://doi.org/10.1016/j.heliyon.2022.e11886
Santoyo G, Urtis-Flores CA, Loeza-Lara PD, Orozco-Mosqueda MD, Glick BR (2021) Rhizosphere colonization determinants by plant growth-promoting rhizobacteria (PGPR). Biology 10:475. https://doi.org/10.3390/biology10060475
Shankar M, Ponraj P, Ilakiam D, Rajendhran J, Gunasekaran P (2012) Genome sequence of the plant growth-promoting bacterium Enterobacter cloacae GS1. J Bacteriol 194:4479–4479. https://doi.org/10.1128/JB.00964-12
Sharma G, Khatri I, Subramanian S (2018) Comparative genomics of myxobacterial chemosensory systems. J Bacteriol 200:e00620–e00617. https://doi.org/10.1128/JB.00620-17
Shilev S (2020) Plant-growth-promoting Bacteria mitigating soil salinity stress in plants. Appl Sci 10:7326. https://doi.org/10.3390/app10207326
Snak A, Vendruscolo ECG, Santos MFd, Fiorini A, Mesa D (2021) Genome sequencing and analysis of plant growth-promoting attributes from Leclercia adecarboxylata. Genet Mol Biology 44:e20200130. https://doi.org/10.1590/1678-4685-gmb-2020-0130
Suarez C, Ratering S, Hain T, Fritzenwanker M, Schnell S (2019) Complete genome sequence of the plant growth-promoting bacterium Hartmannibacter diazotrophicus strain E19T. Int J Genomics. https://doi.org/10.1155/2019/7586430
Tunchai M, Hida A, Oku S, Nakashimada Y, Tajima T, Kato J (2017) Identification and characterization of chemosensors for d-malate, unnatural enantiomer of malate, in Ralstonia pseudosolanacearum. Microbiology 163:233–242. https://doi.org/10.1099/mic.0.000408
Wang H-l, Sun L (2017) Comparative metagenomics reveals insights into the deep-sea adaptation mechanism of the microorganisms in Iheya hydrothermal fields. World J Microbiol Biotechnol 33:86. https://doi.org/10.1007/s11274-017-2255-0
Wang X, Wang C, Li Q, Zhang J, Ji C, Sui J et al (2018) Isolation and characterization of antagonistic bacteria with the potential for biocontrol of soil-borne wheat diseases. J Appl Microbiol 125:1868–1880. https://doi.org/10.1111/jam.14099
Wang X, Zhang M, Loh B, Leptihn S, Ahmed T, Li B (2021) A novel NRPS cluster, acquired by horizontal gene transfer from algae, regulates siderophore iron metabolism in Burkholderia seminalis R456. Int J Biol Macromol 182:838–848. https://doi.org/10.1016/j.ijbiomac.2021.04.051
Yang L, Muhammad I, Chi YX, Wang D, Zhou XB (2022) Straw return and nitrogen fertilization to maize regulate soil properties, microbial community, and enzyme activities under a dual cropping system. Front Microbiol 13:823963. https://doi.org/10.3389/fmicb.2022.823963
Zhang N, Yang DQ, Wang DD, Miao YZ, Shao JH, Zhou X et al (2015) Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genomics 16:685. https://doi.org/10.1186/s12864-015-1825-5
Zuo J, Yin H, Hu J, Miao J, Chen Z, Qi K et al (2019) Lsr operon is associated with AI-2 transfer and pathogenicity in avian pathogenic Escherichia coli. Vet Res 50:109. https://doi.org/10.1186/s13567-019-0725-0
Funding
This work was supported by the Key Research and Development Projects in Heilongjiang, China (GA21B007); and the Outstanding Youth Foundation in Heilongjiang Province, China (JQ2023D001).
Author information
Authors and Affiliations
Contributions
ZW and WX conceived and designed the experiments; WC performed the experiments and analyzed the data; YH participated in the collection of samples and the planning and coordination of the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or nonfinancial interests to disclose.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Xiue Wang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, W., Wang, Z., Xu, W. et al. Genome sequence of Leclercia adecarboxylata QDSM01 with multiple plant growth promoting properties. Plant Growth Regul 102, 445–459 (2024). https://doi.org/10.1007/s10725-023-01071-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-023-01071-4