Abstract
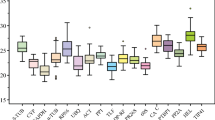
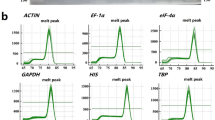
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) is the preferred method for gene expression research, but normalization based on suitable reference genes (RGs) is the key to obtaining reliable gene expression results. In this study, we selected six “commonly used” RGs, two “traditional” housekeeping genes (HKGs), and four novel genes as candidate RGs based on 54 publicly available peel transcriptome libraries that include data from development, bagging, and post-harvest cryopreservation studies of four pear cultivars. The results of this multifaceted assessment from transcriptome and qRT-PCR analyses consistently revealed that ACT6/7/8/9 had the best expression stability among all candidate RGs, and expression of the novel RGs showed greater stability compared with the other “commonly used” RGs and “traditional” HKGs. Among the candidate RGs, ACT6/7/8/9 and Nucleosome Assembly Protein 1 (NAP1) showed superior expression stability and abundance, and they were recommended as the optimal RG combination for gene expression normalization in pear peel. These genome-wide findings provide more reasonable RG usage specifications, and additional useful and reliable RGs as resources for accurate study of gene expression in pear peel studies of different cultivars.




Similar content being viewed by others
Data availability
Genomic and publicly available annotated protein databases for pear (P. bretschneideri) were downloaded from the pear genome project (https://peargenome.njau.edu.cn/). The Arabidopsis data were downloaded from the Arabidopsis Information Resource (https://www.arabidopsis.org/). The gene identity information is listed in the supplemental material (Table S2).
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ (2006) Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6:301–311
Bustin SA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Chen J et al (2015) Identification and testing of reference genes for gene expression analysis in pollen of Pyrus bretschneideri. Sci Hortic 190:43–56
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dai M, Shi Z, Xu C (2015) Genome-wide analysis of sorbitol dehydrogenase (SDH) genes and their differential expression in two sand pear (Pyrus pyrifolia) fruits. Int J Mol Sci 16:13065–13083
Dekkers BJ, Willems L, Bassel GW, van Bolderen-Veldkamp R, Ligterink W, Hilhorst HW, Bentsink L (2012) Identification of reference genes for RT–qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53:28–37
Drøbak B, Franklin-Tong V, Staiger C (2004) The role of the actin cytoskeleton in plant cell signaling. New Phytol 163:13–30
Fu Y, He W, Wang L, Wei Y (2015) Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front Plant Sci 6:475
Gao J, Zhu Y, Zhou W, Molinier J, Dong A, Shen W-H (2012) NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell 24:1437–1447
He X, Wei Y, Kou J, Xu F, Chen Z, Shao X (2018) PpVIN2, an acid invertase gene family member, is sensitive to chilling temperature and affects sucrose metabolism in postharvest peach fruit. Plant Growth Regul 86:169–180
Heng W et al (2018) Characterization and expression analysis of PbEXP genes in the epidermis of pear (Pyrus bretschneideri Rehd.). Plant Growth Regul 84:1–9
Hiwasa K, Rose JK, Nakano R, Inaba A, Kubo Y (2003) Differential expression of seven α-expansin genes during growth and ripening of pear fruit. Physiol Plant 117:564–572
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immunol 6:279–284
Jin X, Fu J, Dai S, Sun Y, Hong Y (2013) Reference gene selection for qPCR analysis in cineraria developing flowers. Sci Hortic 153:64–70
Jung HJ, Park SJ, Kang H (2013) Regulation of RNA metabolism in plant development and stress responses. J Plant Biol 56:123–129
Kang YW, Kim RN, Cho HS, Kim WT, Choi D, Pai H-S (2008) Silencing of a BYPASS1 homolog results in root-independent pleiotrophic developmental defects in Nicotiana benthamiana. Plant Mol Biol 68:423–437
Krzeszowiec W, Rajwa B, Dobrucki J, Gabryś H (2007) Actin cytoskeleton in Arabidopsis thaliana under blue and red light. Biol Cell 99:251–260
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Kwak KJ, Kim YO, Kang H (2005) Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J Exp Bot 56:3007–3016
Kwon SJ, Choi EY, Choi YJ, Ahn JH, Park OK (2006) Proteomics studies of post-translational modifications in plants. J Exp Bot 57:1547–1551
Lankenau S, Barnickel T, Marhold J, Lyko F, Mechler BM, Lankenau D-H (2003) Knockout targeting of the Drosophila Nap1 gene and examination of DNA repair tracts in the recombination products. Genetics 163:611–623
Li Q-F, Sun SS, Yuan D-Y, Yu H-X, Gu M-H, Liu Q-Q (2010) Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep 28:49–57
Liu Z, Zhu Y, Gao J, Yu F, Dong A, Shen WH (2009) Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J 59:27–38
Liu P et al (2013) Molecular analysis of the processes of surface brown spot (SBS) formation in pear fruit (Pyrus bretschneideri Rehd. Cv. Dangshansuli) by de novo transcriptome assembly. PLoS ONE 8:e74217
Liu B, Wang L, Wang S, Li W, Liu D, Guo X, Qu B (2019) Transcriptomic analysis of bagging-treated ‘Pingguo’pear shows that MYB4-like1, MYB4-like2, MYB1R1 and WDR involved in anthocyanin biosynthesis are up-regulated in fruit peels in response to light. Sci Hortic 244:428–434
Long X-Y et al (2010) Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol Biol 74:307–311
Meagher RB, McKinney EC, Kandasamy M (1999) Isovariant dynamics expand and buffer the responses of complex systems: the diverse plant actin gene family. Plant Cell 11:995–1005
Müller OA, Grau J, Thieme S, Prochaska H, Adlung N, Sorgatz A, Bonas U (2015) Genome-wide identification and validation of reference genes in infected tomato leaves for quantitative RT-PCR analyses. PLoS ONE 10:e0136499
Nham NT et al (2015) A transcriptome approach towards understanding the development of ripening capacity in ‘Bartlett’pears (Pyrus communis L.). BMC Genomics 16:762
Nham NT, Macnish AJ, Zakharov F, Mitcham EJ (2017) ‘Bartlett’pear fruit (Pyrus communis L.) ripening regulation by low temperatures involves genes associated with jasmonic acid, cold response, and transcription factors. Plant Sci 260:8–18
Ni J, Bai S, Gao L, Qian M, Zhong L, Teng Y (2017) Identification, classification, and transcription profiles of the B-type response regulator family in pear. PLoS ONE 12:e0171523
Park Y-J, Luger K (2006) Structure and function of nucleosome assembly proteins. Biochem Cell Biol 84:549–549
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat Protoc 11:1650–1667
Pfaffl MW (2004) Quantification strategies in real-time PCR. AZ Quant PCR 1:89–113
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Pierantoni L, Dondini L, De Franceschi P, Musacchi S, Winkel BS, Sansavini S (2010) Mapping of an anthocyanin-regulating MYB transcription factor and its expression in red and green pear, Pyrus communis. Plant Physiol Biochem 48:1020–1026
Pihur V, Datta S, Datta S (2009) RankAggreg, an R package for weighted rank aggregation. BMC Bioinf 10:62
Ramakers C, Ruijter JM, Deprez RHL, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Sieburth LE, Lee DK (2010) BYPASS1: How a tiny mutant tells a big story about root-to-shoot signaling. J Integr Plant Biol 52:77–85
Silverman IM, Li F, Gregory BD (2013) Genomic era analyses of RNA secondary structure and RNA-binding proteins reveal their significance to post-transcriptional regulation in plants. Plant Sci 205:55–62
Thompson JD, Gibson TJ, Higgins DG (2003) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinf 1:1–22
Tripurani SK, Nakaminami K, Thompson KB, Crowell SV, Guy CL, Karlson DT (2011) Spatial and temporal expression of cold-responsive DEAD-box RNA helicases reveals their functional roles during embryogenesis in Arabidopsis thaliana. Plant Mol Biol Rep 29:761–768
Van Norman JM, Sieburth LE (2007) Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J 49:619–628
Van Norman JM, Frederick RL, Sieburth LE (2004) BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol 14:1739–1746
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(research0034):0031
Vembar SS, Macpherson CR, Sismeiro O, Coppée J-Y, Scherf A (2015) The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages. Genome Biol 16:212
Vierstra RD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397
Wan H et al (2011) Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem Biophys Res Commun 416:24–30
Wang Y, Zhang W-Z, Song L-F, Zou J-J, Su Z, Wu W-H (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148:1201–1211
Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013) The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol 162:885–896
Wang R et al (2017a) Genome-wide identification of the MADS-box transcription factor family in pear (Pyrus bretschneideri) reveals evolution and functional divergence. Peer Journal 5:e3776
Wang Y, Zhang X, Wang R, Bai Y, Liu C, Yuan Y, Yang Y, Yang S (2017b) Differential gene expression analysis of ‘Chili’(Pyrus bretschneideri) fruit pericarp with two types of bagging treatments. Hortic Res 4:17005
Wang Z, Du H, Zhai R, Song L, Ma F, Xu L (2017c) Transcriptome analysis reveals candidate genes related to color fading of ‘Red Bartlett’(Pyrus communis L.). Front Plant Sci 8:455
Wen R et al (2014) UBC 13, an E2 enzyme for L ys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J 80:424–436
Wilhelm BT, Landry J-R (2009) RNA-Seq—quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48:249–257
Wu T, Zhang R, Gu C, Wu J, Wan H, Zhang S, Zhang S (2012) Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr J Agric Res 7:3701–3704
Wu J et al (2013a) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Wu J et al (2013b) Identification of differentially expressed genes related to coloration in red/green mutant pear (Pyrus communis L.). Tree Genet Genomes 9:75–83
Wu J et al (2018) Diversification and independent domestication of Asian and European pears. Genome Biol 19:77
Xie F, Xiao P, Chen D, Xu L, Zhang B (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80:75–84
Xu L, Ménard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH (2009) The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J 57:279–288
Xue C et al (2019) PbrMYB169 positively regulates lignification of stone cells in pear fruit. J Exp Bot 70:1801–1814
Yang Y, Yao G, Yue W, Zhang S, Wu J (2015) Transcriptome profiling reveals differential gene expression in proanthocyanidin biosynthesis associated with red/green skin color mutant of pear (Pyrus communis L.). FrontPlant Sci 6:795
Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10:755–764
Zhang D, Yu B, Bai J, Qian M, Shu Q, Su J, Teng Y (2012) Effects of high temperatures on UV-B/visible irradiation induced postharvest anthocyanin accumulation in ‘Yunhongli No. 1’(Pyrus pyrifolia Nakai) pears. Sci Hortic 134:53–59
Zhu Y, Dong A, Meyer D, Pichon O, Renou J-P, Cao K, Shen W-H (2006) Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18:2879–2892
Acknowledgements
This work was supported by Natural Science Foundation of Fujian Province, China (2018J01702); the National Natural Science Foundation of China (31801814); the Fundamental Research Funds for the Fujian Universities (JK2017017); and the Central Government Guides Local Science and Technology Development Projects (2017L3001). We thank Mallory Eckstut, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
CJQ, CQX, and CCM conceived and designed the experiments, analyzed the data, and wrote the manuscript; CCM, WTY, and CZM performed the experiments; XGF and CZY contributed plant materials; YYF and WW contributed analytical tools; XQ and GXY helped analyze data; CJQ revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors of this manuscript declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, C., Wang, T., Cai, Z. et al. Genome-wide identification and validation of optimal reference genes for gene expression normalization in pear peel. Plant Growth Regul 92, 249–261 (2020). https://doi.org/10.1007/s10725-020-00634-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00634-z