Abstract
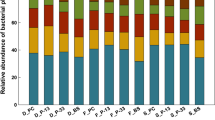
Assessing the effects of transgenic crops on rhizobacterial communities has catalyzed numerous studies in the recent past decades. To address further to this concern, we designed research to examine the variations in the bacterial diversity profiles of transgenic insect-resistant maize line 2A-5 (2A5) carrying mcry1Ab and mcry2Ab genes and its control Zheng58 (Z58), at different developmental stages under natural field conditions. High-throughput sequencing of the 16S rDNA gene (V3–V4) hyper-variable region via Illumina MiSeq revealed no significant shifts in the overall α and β-diversity. However, some developmental stage-dependent discrepancies were observed in the relative abundances of microbial communities. At the jointing stage, members of the phylum Proteobacteria, and the genera Rahnella, Kaistobacter, Pseudomonas and Arthrobacter were greatly enriched in the rhizosphere soil of 2A5 and Z58 compared to bulk, and surrounding soil while de-riched with the microbes belonging to the genera Bacillus and Flavobacterium. In addition, the relative abundances of nitrogen fixing microbes i.e. Arthrobacter, Burkholderia and Pseudomonas in the rhizosphere samples of 2A5 showed exuberance compared to those of Z58 at the flowering and maturation stages, implying that genetic modification might play a role in the enrichment/derichment of some nitrogen fixing microbes. The results of our study indicate little to no impacts of transgenic maize carrying mcry1Ab and mcry2Ab genes and also, the changes in the rhizosphere bacterial communities are imposed by host plant during different developmental stages.






Similar content being viewed by others
References
Abarenkov K, Nilsson RH, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vralstad T, Liimatainen K, Peintner U, Koljalg U (2010) The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol 186(2):281–285. https://doi.org/10.1111/j.1469-8137.2009.03160.x
Ahasan MS, Picard J, Elliott L, Kinobe R, Owens L, Ariel E (2017) Evidence of antibiotic resistance in Enterobacteriales isolated from green sea turtles, Chelonia mydas on the Great Barrier Reef. Mar Pollut Bull 120(1–2):18–27. https://doi.org/10.1016/j.marpolbul.2017.04.046
Bakhsh A, Khabbazi SD, Baloch FS, Demirel U, Caliskan ME, Hatipoglu R, Ozcan S, Ozkan H (2015) Insect-resistant transgenic crops: retrospect and challenges. Turk J Agric For 39(4):531–548. https://doi.org/10.3906/tar-1408-69
Bates SL, Zhao JZ, Roush RT, Shelton AM (2005) Insect resistance management in GM crops: past, present and future. Nat Biotechnol 23(1):57–62. https://doi.org/10.1038/nbt1056
Bever JD, Platt TG, Morton ER (2012) Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu Rev Microbiol 66:265–283. https://doi.org/10.1146/annurev-micro-092611-150107
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488(7409):91–95. https://doi.org/10.1038/nature11336
Bulgarelli D, Garrido-Oter R, Munch PC, Weiman A, Droge J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17(3):392–403. https://doi.org/10.1016/j.chom.2015.01.011
Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T (2011) Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA 108(34):14288–14293. https://doi.org/10.1073/pnas.1101591108
Carpenter JE (2010) Peer-reviewed surveys indicate positive impact of commercialized GM crops. Nat Biotechnol 28(4):319–321. https://doi.org/10.1038/nbt0410-319
Chauhan PS, Chaudhry V, Mishra S, Nautiyal CS (2011) Uncultured bacterial diversity in tropical maize (Zea mays L.) rhizosphere. J Basic Microbiol 51(1):15–32. https://doi.org/10.1002/jobm.201000171
Corbin DR, Romano CP (2010) U.S. Patent No. 7,700,830. Washington, DC: U.S. Patent and Trademark Office
Dale PJ, Clarke B, Fontes EM (2002) Potential for the environmental impact of transgenic crops. Nat Biotechnol 20(6):567–574. https://doi.org/10.1038/nbt0602-567
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996. https://doi.org/10.1038/Nmeth.2604
Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA 112(8):E911–E920. https://doi.org/10.1073/pnas.1414592112
Fernandez-Gonzalez AJ, Lasa AV, Fernandez-Lopez M (2018) Whole-genome sequences of two Arthrobacter strains isolated from a holm oak rhizosphere affected by wildfire. Microbiol Resour Ann 6(11):e00071-18
Firakova S, Sturdikova M, Muckova M (2007) Bioactive secondary metabolites produced by microorganisms associated with plants. Biologia 62(3):251–257. https://doi.org/10.2478/s11756-007-0044-1
Gatehouse JA (2008) Biotechnological prospects for engineering insect-resistant plants. Plant Physiol 146(3):881–887. https://doi.org/10.1104/pp.107.111096
Gyaneshwar P, Hirsch AM, Moulin L, Chen WM, Elliott GN, Bontemps C, Estrada-de los Santos P, Gross E, dos Reis FB, Sprent JI, Young JPW, James EK (2011) Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol Plant Microbe Interact 24(11):1276–1288. https://doi.org/10.1094/Mpmi-06-11-0172
Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60(4):579–598. https://doi.org/10.1007/s13213-010-0117-1
Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ (2008) Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol Ecol 65(2):193–201. https://doi.org/10.1111/j.1574-6941.2008.00535.x
Hu LF, Robert CAM, Cadot S, Zhang X, Ye M, Li BB, Manzo D, Chervet N, Steinger T, van der Heijden MGA, Schlaeppi K, Erb M (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9:1–13. https://doi.org/10.1038/s41467-018-05122-7
Inceoglu O, Salles JF, van Overbeek L, van Elsas JD (2010) Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl Environ Microbiol 76(11):3675–3684. https://doi.org/10.1128/Aem.00040-10
Khan GA, Bakhsh A, Ghazanfar M, Riazuddin S, Husnain T (2013) Development of transgenic cotton lines harboring a pesticidal gene (cry1Ab). Emir J Food Agric 25(6):434–442. https://doi.org/10.9755/ejfa.v25i6.13133
Koul B, Srivastava S, Sanyal I, Tripathi B, Sharma V, Amla DV (2014) Transgenic tomato line expressing modified Bacillus thuringiensis cry1Ab gene showing complete resistance to two lepidopteran pests. Springerplus 3:84. https://doi.org/10.1186/2193-1801-3-84
Lai JS, Zhao HM, Song WB, Cui Y (2015) Anti insect gene mCry2Ab and its application. China Patent No. 2014104831432
Li XZ, Rui JP, Mao YJ, Yannarell A, Mackie R (2014) Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem 68:392–401. https://doi.org/10.1016/j.soilbio.2013.10.017
Li ZL, Bu NS, Chen XP, Cui J, Xiao MQ, Song ZP, Nie M, Fang CM (2018) Soil incubation studies with Cry1Ac protein indicate no adverse effect of Bt crops on soil microbial communities. Ecotox Environ Saf 152:33–41. https://doi.org/10.1016/j.ecoenv.2017.12.054
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71(12):8228–8235. https://doi.org/10.1128/Aem.71.12.8228-8235.2005
Lu GH, Tang CY, Hua XM, Cheng J, Wang GH, Zhu YL, Zhang LY, Shou HX, Qi JL, Yang YH (2018) Effects of an EPSPS-transgenic soybean line ZUTS31 on root-associated bacterial communities during field growth. PLoS ONE 13(2):e0192008. https://doi.org/10.1371/journal.pone.0192008
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Moreira FD, da Costa PB, de Souza R, Beneduzi A, Lisboa BB, Vargas LK, Passaglia LMP (2016) Functional abilities of cultivable plant growth promoting bacteria associated with wheat (Triticum aestivum L.) crops. Genet Mol Biol 39(1):111–121. https://doi.org/10.1590/1678-4685-Gmb-2015-0140
Najjuka CF, Kateete DP, Kajumbula HM, Joloba ML, Essack SY (2016) Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes 9:235. https://doi.org/10.1186/s13104-016-2049-8
Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D (2014) Niche and host-associated functional signatures of the root surface microbiome. Nat Commun 5:1–9. https://doi.org/10.1038/ncomms5950
Ouyang B, Gu XF, Holford P (2017) Plant genetic engineering and biotechnology: a sustainable solution for future food security and industry. Plant Growth Regul 83(2):171–173. https://doi.org/10.1007/s10725-017-0300-5
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110(16):6548–6553. https://doi.org/10.1073/pnas.1302837110
Peng XX, Guo F, Ju F, Zhang T (2014) Shifts in the microbial community, nitrifiers and denitrifiers in the biofilm in a full-scale rotating biological contactor. Environ Sci Technol 48(14):8044–8052. https://doi.org/10.1021/es5017087
Pimentel DS, Raven PH (2000) Bt corn pollen impacts on nontarget Lepidoptera: assessment of effects in nature. Proc Natl Acad Sci USA 97(15):8198–8199. https://doi.org/10.1073/pnas.97.15.8198
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–D596. https://doi.org/10.1093/nar/gks1219
Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N (2010) The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12(11):2885–2893. https://doi.org/10.1111/j.1462-2920.2010.02258.x
Singh AK, Dubey SK (2016) Current trends in Bt crops and their fate on associated microbial community dynamics: a review. Protoplasma 253(3):663–681. https://doi.org/10.1007/s00709-015-0903-5
Singh RJ, Ahlawat IPS, Singh S (2013) Effects of transgenic Bt cotton on soil fertility and biology under field conditions in subtropical inceptisol. Environ Monit Assess 185(1):485–495. https://doi.org/10.1007/s10661-012-2569-1
Song FS, Ni DH, Li Hao, Duan Y, Yang YC, Ni JL, Lu XZ, Wei PC, Li L, Yang J (2014) A novel synthetic Cry1Ab gene resists rice insect pests. Genet Mol Res 13:2394–2408. https://doi.org/10.4238/2014.April.3.12
Strain KE, Lydy MJ (2015) The fate and transport of the Cry1Ab protein in an agricultural field and laboratory aquatic microcosms. Chemosphere 132:94–100. https://doi.org/10.1016/j.chemosphere.2015.03.005
Umbeck P, Johnson G, Barton K, Swain W (1987) Genetically transformed cotton (Gossypium-Hirsutum-L.) plants. Biotechnology 5(3):263–266. https://doi.org/10.1038/nbt0387-263
Verbruggen E, Kuramae EE, Hillekens R, de Hollander M, Kiers ET, Roling WFM, Kowalchuk GA, van der Heijden MGA (2012) Testing potential effects of maize expressing the Bacillus thuringiensis Cry1Ab endotoxin (Bt maize) on mycorrhizal fungal communities via DNA- and RNA-based pyrosequencing and molecular fingerprinting. Appl Environ Microbiol 78(20):7384–7392. https://doi.org/10.1128/Aem.01372-12
Wagner MR, Lundberg DS, Coleman-Derr D, Tringe SG, Dangl JL, Mitchell-Olds T (2014) Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecol Lett 17(6):717–726. https://doi.org/10.1111/ele.12276
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. https://doi.org/10.1128/Aem.00062-07
Wang JY, Wu LK, Tantai HP, Khan MU, Letuma P, Wu HM, Zhang SK, Chen T, Lin S, Lin WX (2019) Properties of bacterial community in the rhizosphere soils of Achyranthes bidentata tolerant to consecutive monoculture. Plant Growth Regul 89(2):167–178. https://doi.org/10.1007/s10725-019-00523-0
Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vazquez-Baeza Y, Birmingham A, Hyde ER, Knight R (2017) Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5(1):27. https://doi.org/10.1186/s40168-017-0237-y
Wen ZL, Yang MK, Du MH, Zhong ZZ, Lu YT, Wang GH, Hua XM, Fazal A, Mu CH, Yan SF, Zhen Y, Yang RW, Qi JL, Hong Z, Lu GH, Yang YH (2019) Enrichments/derichments of root-associated bacteria related to plant growth and nutrition caused by the growth of an EPSPS-transgenic maize line in the field. Front Microbiol 10:1335. https://doi.org/10.3389/fmicb.2019.01335
Yan Y, Kuramae EE, de Hollander M, Klinkhamer PGL, van Veen JA (2017) Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J 11(1):56–66. https://doi.org/10.1038/ismej.2016.108
Yoneyama K, Xie X, Yoneyama K (2011) Strigolactones as chemical signals for plant-plant and plant-microbe interactions in the rhizosphere. Phytopathology 101(6):S233–S233
Yuan J, Chaparro JM, Manter DK, Zhang RF, Vivanco JM, Shen QR (2015) Roots from distinct plant developmental stages are capable of rapidly selecting their own microbiome without the influence of environmental and soil edaphic factors. Soil Biol Biochem 89:206–209. https://doi.org/10.1016/j.soilbio.2015.07.009
Zhou JZ, Wu LY, Deng Y, Zhi XY, Jiang YH, Tu QC, Xie JP, Van Nostrand JD, He ZL, Yang YF (2011) Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5(8):1303–1313. https://doi.org/10.1038/ismej.2011.11
Acknowledgements
We are grateful to Prof. Xuehui Dong of College of Agronomy and Biotechnology, China Agriculture University, for providing maize seeds of transgenic insect-resistant maize 2A5 and its control variety Z58. This work was supported by the grants from National Important Science & Technology Specific Project (2016ZX08011-003), the National Natural Science Foundation of China (31870495), and the Program for Changjiang Scholars and Innovative Research Team in University from the Ministry of Education of China (IRT_14R27)
Author information
Authors and Affiliations
Contributions
YHY, GHL, and JLQ conceived and designed the experiments; AF, ZLW, YTL, and MKY performed the experiments; AF, ZLW, XMH, and YTL did data analysis; XMH, TMY, HWH, HYL, and XMW contributed to the resources; AF, and ZLW wrote the draft of manuscript; YHY, GHL, and JLQ contributed to reviewing and editing the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no potential conflict of interest. The funding sponsors had no role in the design of the study, collection, analyses, interpretation of the data, writing of the manuscript, and in the decision to publish the results.
Ethical approval
The Ministry of Agriculture of the People's Republic of China issued permission for the location. The field studies did not involve endangered species. The experimental field was not privately owned in any way.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fazal, A., Wen, ZL., Lu, YT. et al. Assembly and shifts of the bacterial rhizobiome of field grown transgenic maize line carrying mcry1Ab and mcry2Ab genes at different developmental stages. Plant Growth Regul 91, 113–126 (2020). https://doi.org/10.1007/s10725-020-00591-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00591-7