Abstract
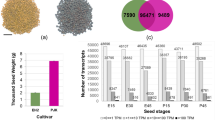
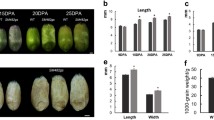
Seed size is one of the most important determinants of yield in lotus (Nelumbo nucifera). The cotyledon, which is responsible for nutrient storage in the mature seed, is the major factor affecting seed size in this economically important crop. Here, transcriptome analysis on cotyledons were performed during the rapid expansion stage of two lotus cultivars with different seed size and yield, China Antique and Jianxuan-17, at 9, 12, and 15 days after pollination (DAP). We identified 22,549 genes, including 2414 novel genes. Among them, 8437 genes were differentially expressed between CA and JX from 9 to 15 DAP. Gene ontology analysis suggested that these DEGs were significantly enriched in cell proliferation and gene expression. Dozens of DEGs are involved in brassinosteroids (BRs) biosynthesis and signaling pathway. Nine genes controlling seed size by cell number and size were candidate genes regulating lotus seed size. There was a notable difference in the expression patterns and level of starch-synthesis genes between two cultivars. NNU_20629 and NNU_05331 were likely responsible for the difference in starch accumulation between CA and JX, which might lead to their different yield. Pairwise comparisons of our transcriptome data provide insights into lotus seed development, which could facilitate projects aimed at breeding lotus with improved traits.







Similar content being viewed by others
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Cheng L, Liu X, Yin JJ, Yang JQ, Li Y, Hui LC, Li SY, LI LJ (2016) Activity and expression of ADP-glucose pyrophosphorylase during rhizome formation in lotus (Nelumbo nucifera Gaertn.). Bot Stud 57:26–36
Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26:573–582
Deng YY, Li JQ, Wu SF, Zhu YP, Chen YW, He FC (2006) Integrated nr database in protein annotation system and its localization. Comput Eng 32:71–74
Divi UK, Krishna P (2009) Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol 26:131–136
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Finn RD, Bateman A, Clements J, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2013) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230
Folsom JJ, Begcy K, Hao X, Wang D, Walia H (2014) Rice fertilization-independent endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol 165:238–248
Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17:52–60
Huang HY, Jiang WB, Hu YW, Wu P, Zhu JY, Liang WQ, Wang ZY, Lin WH (2013) BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol Plant 6:456–469
Jiang WB, Lin WH (2013) Brassinosteroid functions in Arabidopsis seed development. Plant Signal Behav 8:e25928
Jiang HY, Zhang J, Wang JM, Xia M, Zhu SW, Cheng BJ (2013a) RNA interference-mediated silencing of the starch branching enzyme gene improves amylose content in rice. Genet Mol Res 12:2800–2808
Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH (2013b) Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol 162:1965–1977
Jofuku KD, Omidyar PK, Gee Z, Okamuro JK (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci USA 102:3117–3122
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280
Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43:561–571
Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, He Y, Zhang Q (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269
Li Z, Tang L, Qiu J, Zhang W, Wang Y, Tong X, Wei X, Hou Y, Zhang J (2016) Serine carboxypeptidase 46 regulates grain filling and seed germination in rice (Oryza sativa L.). PLoS ONE 11:e0159737
Lou Q, Liu Y, Qi Y, Jiao S, Tian F, Jiang L, Wang Y (2014) Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J Exp Bot 65:3157–3164
Manoli A, Trevisan S, Quaggiotti S, Varotto S (2018) Identification and characterization of the BZR transcription factor family and its expression in response to abiotic stresses in Zea mays L. Plant Growth Regul 84:423–436
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Ming R, VanBuren R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M, Li J, Bowers JE, Tang H, Lyons E, Ferguson AA, Narzisi G, Nelson DR, Blaby-Haas CE, Gschwend AR, Jiao Y, Der JP, Zeng F, Han J, Min XJ, Hudson KA, Singh R, Grennan AK, Karpowicz SJ, Watling JR, Ito K, Robinson SA, Hudson ME, Yu Q, Mockler TC, Carroll A, Zheng Y, Sunkar R, Jia R, Chen N, Arro J, Wai CM, Wafula E, Spence A, Han Y, Xu L, Zhang J, Peery R, Haus MJ, Xiong W, Walsh JA, Wu J, Wang ML, Zhu YJ, Paull RE, Britt AB, Du C, Downie SR, Schuler MA, Michael TP, Long SP, Ort DR, Schopf JW, Gang DR, Jiang N, Yandell M, dePamphilis CW, Merchant SS, Paterson AH, Buchanan BB, Li S, Shen-Miller J (2013) Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol 14:R41
Nakagawa H, Tanaka A, Tanabata T, Ohtake M, Fujioka S, Nakamura H, Ichikawa H, Mori M (2011) SHORT GRAIN1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol 158:1208–1219
Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102:3123–3128
Regina A, Kosar-Hashemi B, Ling S, Li Z, Rahman S, Morell M (2010) Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J Exp Bot 61:1469–1482
Sarma K, Sen P, Barooah M, Choudhury MD, Roychoudhury S, Modi MK (2014) Structural comparison, substrate specificity, and inhibitor binding of AGPase small subunit from monocot and dicot: present insight and future potential. Biomed Res Int 2014:1–20
Savadi S (2018) Molecular regulation of seed development and strategies for engineering seed size in crop plants. Plant Growth Regul 84:401–422
Savolainen V, Chase MW (2003) A decade of progress in plant molecular phylogenetics. Trends Genet 19:717–724
Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133:251–261
Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ (2002) Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci USA 99:1724–1729
Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, Sessa G (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J 74:905–919
Sun X, Shantharaj D, Kang X, Ni M (2010) Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr Opin Plant Biol 13:611–620
Syahariza ZA, Sar S, Hasjim J, Tizzotti MJ, Gilbert RG (2013) The importance of amylose and amylopectin fine structures for starch digestibility in cooked rice grains. Food Chem 136:742–749
Toyosawa Y, Kawagoe Y, Matsushima R, Crofts N, Ogawa M, Fukuda M, Kumamaru T, Okazaki Y, Kusano M, Saito K, Toyooka K, Sato M, Ai Y, Jane JL, Nakamura Y, Fujita N (2016) Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol 170:1255–1270
Tuncel A, Okita TW (2013) Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: expectations and unanticipated outcomes. Plant Sci 211:52–60
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Wang L, Fu J, Li M, Fragner L, Weckwerth W, Yang P (2016) Metabolomic and proteomic profiles reveal the dynamics of primary metabolism during seed development of lotus (Nelumbo nucifera). Front Plant Sci 7:750
Wu Y, Fu Y, Zhao S, Gu P, Zhu Z, Sun C, Tan L (2016) CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J 14:377–386
Xie C, Mao X, Huang J, Ding Y, Wu JM, Dong S, Kong L, Gao G, Li CY, Wei LP (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322
Xu C, Liu Y, Li Y, Xu X, Xu C, Li X, Xiao J, Zhang Q (2015) Differential expression of GS5 regulates grain size in rice. J Exp Bot 66:2611–2623
Yang M, Zhu L, Pan C, Xu L, Liu Y, Ke W, Yang P (2015) Transcriptomic analysis of the regulation of rhizome formation in temperate and tropical lotus (Nelumbo nucifera). Sci Rep 5:13059
Yang M, Zhu L, Li L, Li J, Xu L, Feng J, Liu Y (2017) Digital gene expression analysis provides insight into the transcript profile of the genes involved in aporphine alkaloid biosynthesis in lotus (Nelumbo nucifera). Front Plant Sci 8:80
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14
Zhang Y, Liang W, Shi J, Xu J, Zhang D (2013) MYB56 encoding a R2R3 MYB transcription factor regulates seed size in Arabidopsis thaliana. J Integr Plant Biol 55:1166–1178
Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J (2002) Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol 130:1221–1229
Zhu JY, Sae-Seaw J, Wang ZY (2013) Brassinosteroid signalling. Development 140:1615–1620
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 31700197) and Key Research Program of Frontier Sciences, CAS, Grant No. QYZDB-SSW-SMC017.
Author information
Authors and Affiliations
Contributions
JL performed the experiment and wrote the paper. TS and LH analyzed the data. DH and TMN conducted some experiments. PY designed the experiment and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J., Shi, T., Huang, L. et al. Systematic transcriptomic analysis provides insights into lotus (Nelumbo nucifera) seed development. Plant Growth Regul 86, 339–350 (2018). https://doi.org/10.1007/s10725-018-0433-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0433-1