Abstract
The three-amino-acid-loop-extension (TALE) proteins are a class of transcription factors that regulate meristem differentiation during plant growth and development. In this study, we identified a total of 45 members of the tobacco TALE (NtTALE) transcription factor family. Based on domain type, the NtTALE genes were divided into BELL subfamily and KNOX subfamily. Cis-acting element analysis showed that the NtTALE gene promoters had a variety of response elements, including light-responsive elements, hormonal and abiotic stresses responsive elements, plant development related elements. Among the 45 NtTALE genes, the expression patterns of various plant tissues at different stages were different, 10 responded to alkali stress, 19 responded to dehydration treatment, 14 responded to cold stress, two responded to salt stress, and 17 responded to Ralstonia solanacearum infection. Phylogenetic analysis showed that the number of TALE genes in Nicotiana was significantly expanded compared with that in other Solanaceae. The KNOXIII subfamily members (NtTALE29, NtTALE45) were found only in Nicotiana tabacum. This study provides a theoretical basis for further research on the biological function of NtTALE genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Homeobox (HB) gene is a class of genes which have a 60 amino acid helix-turn-helix DNA binding motif. The characteristic domain is called a Homeodomain (HD) (Desplan et al. 1988). TALE transcription factors are a special class of HB genes. It encode one atypical HD consisting of 63 amino acids, with an additional 3 amino acid (PYP, proline-tyrosine-proline) residues linked between the first and second helix regions (Bertolino et al. 1995).
The TALE gene family consists of the KNOX subfamily and the BELL subfamily. The KNOX subfamily includes four domains: KNOX1, KNOX2, ELK, and Homeodomain. KNOX proteins can be divided into three types: KNOXI, KNOXII, and KNOXIII. KNOXI genes have been found to be mainly expressed in meristem and play an important roles in plant growth and development (Hay and Tsiantis 2010). For example, in Arabidopsis thaliana (A. thaliana), the SHOOT-MERISTEMLESS (STM) gene encoding KNOXI protein was found to be necessary for the formation of shoot apical meristem (SAM) during embryogenesis (Long et al. 1996). KNOXII genes can regulate the formation of plant secondary walls (Li et al. 2012). KNOXIII genes are currently only found in dicots, a novel KNOX gene that lacks the HD. KNATM found in A. thaliana was expressed in the proximal lateral domain of organ primordium and the boundary of mature organs (Magnani and Hake 2008). The BELL subfamily, also known as the BLH subfamily, includes POX, HD domains. The POX domain, also known as the MID domain, which is composed of two domains: SKY and BELL. The BELL subfamily plays an important role in the response to stress, the development of meristems, hormone biosynthesis and signal transduction (Niu and Fu 2022). In previous research on A. thaliana, BELL1 was found to be associated with ovule development (Brambilla et al. 2007). ATH1 has been shown to regulate the growth of the interface between stems, meristem and organ primordium (Gomez-Mena and Sablowski 2008). BLH1 and KNAT3 have been shown to regulate seed germination and early seedling development by directly regulating ABI3 expression (Kim et al. 2013). ABI3 mediates both plant development and the stress response (Xu and Cai 2019). SlBEL11 plays an important role in chloroplast development and chlorophyll synthesis in tomato fruit (Meng et al. 2018).
TALE transcription factors are widely found in plants and have been identified in a variety of plants such as Punica granatum (Wang et al. 2020), Prunus mume (Yang et al. 2022), Gossypium arboretum (Ma et al. 2019), Phyllostachys edulis (Xu et al. 2019), Ananas comosus (Ali et al. 2019). There have been many relevant investigations proving it plays an important role in the growth and development of plants and the response to the eliminate external environment (Zhao et al. 2019; Li et al. 2022; Hou et al. 2021). In previous research, TALE has been found to be related to various biological processes such as the maintenance of organ morphology, hormone regulation and tuber formation (Belles-Boix et al. 2006; Shani et al. 2006; Kondhare et al. 2019). TALE proteins were found in A. thaliana to control meristem formation and/or maintenance, organ position, and several aspects of reproductive phase. And they are also involved in the regulation of several hormonal pathways, providing a link between gene regulatory networks and SAM signaling (Hamant and Pautot 2010). Research has shown that TALE genes plays an important role in fruit quality and maturity of many fruit crops (Brian et al. 2021; Costa et al. 2020; Shahan et al. 2019).
Tobacco (Nicotiana tabacum L.) is a cash crop that harvests leaves. Tobacco is also an important model plant" (Bally et al. 2018). Functional verification research of many plant genes is carried out in tobacco (Li et al. 2023; Chen et al. 2023; Zhao et al. 2023). With global warming and frequent natural disasters, tobacco is facing and suffering from many stresses. TALE gene encode is a transcription factor that can participate in the maintenance of organ morphology and respond to a variety of stresses. The study of TALE genes is of great significance for tobacco stress resistance and growth and development. The completion of tobacco genome sequencing has greatly facilitated the study of gene function in tobacco (Sierro et al. 2014). In this study, the physicochemical properties, subcellular localization, signal peptides, conserved motifs, gene structure, cis-acting elements, protein secondary and 3D structure, protein–protein interaction networks (PPI) analysis, gene expression patterns and evidence of expansion in the Solanaceae of NtTALE genes were analyzed. This study provides a theoretical basis for further revealing the role of TALE transcription factors in tobacco.
Materials and methods
Identification of the TALE genes: To identify the TALE genes, A. thaliana TALE (AtTALE) protein sequences, Nicotiana tabacum cv. TN90 and Solanum ptychanthum genomic data were downloaded from the TAIR database (https://www.arabidopsis.org/, accessed on 27 March 2023) (Lamesch et al. 2012), the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 27 March 2023) (Sayers et al. 2022) and Solanaceae Genomics Network (https://solgenomics.net/, accessed on 7 March 2024) (Fernandez-Pozo et al. 2015), respectively. TBtools was used to blast the protein sequence of AtTALE transcription factor family proteins in tobacco and Solanum ptychanthum, respectively, and the parameters were all default (Chen et al. 2023). The domains of candidate NtTALE genes were identified using the CDD database (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml, accessed on 8 March 2024) (Lu et al. 2020). When there were multiple transcripts of the same gene, the longest transcript was selected as the TALE gene.
Phylogenetic and orthologous gene analysis of TALE genes: Nicotiana sylvestris, Nicotiana tomentosiformis, Solanum lycopersicum, Capsicum annuum, Nicotiana benthamiana, Oryza sativa subsp. Japonica, Oryza sativa subsp. Indica, Solanum melongena, Solanum pimpinellifolium, Solanum pennellii, and Solanum tuberosum TALE protein sequences were downloaded from Plant Transcription Factor Database (https://planttfdb.gao-lab.org/, accessed on 7 March 2024) (Tian et al. 2019). The TALE protein sequences were aligned by ClustalW. The phylogenetic tree was build by MEGA7.0 (Kumar et al. 2016). The NJ method was used, with the parameters were set to Bootstrap method 1000 times, poisson model, and pairwise deleetion. The phylogenetic tree was beautified by the iTOL online website (https://itol.embl.de/, accessed on 15 March 2024) (Letunic and Bork 2021). The GmBLH4 protein sequence was downloaded from Phytozome database (https://phytozome-next.jgi.doe.gov/, accessed on 18 April 2024) (Goodstein et al. 2012). Orthologous genes from Capsicum annuum, Solanum lycopersicum, Solanum melongena, Solanum tuberosum, Glycine max, Nicotiana benthamiana, Nicotiana sylvestris, Nicotiana tomentosiformis and Nicotiana tabacum were analyzed by OrthoFinder (Emms and Kelly 2019).
Physicochemical properties, subcellular localization, and signaling peptide of NtTALE genes. The AA, MW, pI, AI, and GRAVY of the NtTALE genes were predicted by the ExPASy ProtPara online website (https://web.expasy.org/protparam/, accessed on 30 March 2023) (Duvaud et al. 2021). The subcellular localization of NtTALE genes were predicted by the online website WoLF PSORT(https://wolfpsort.hgc.jp/, accessed on 30 March 2023) (Horton et al. 2007). The signaling peptides of the NtTALE genes were predicted by the online website SignalP 5.0 (https://services.healthtech.dtu.dk/services/Signal-P-5.0/, accessed on 7 April 2023) (Nielsen et al. 1999).
Conservation motif and gene structure of the NtTALE genes. The conserved motifs of the NtTALE proteins were analyzed by the online website MEME (http://meme-suite.org, accessed on 31 March 2023) (Bailey et al. 2015). The maximum number of motifs was set to 10, and the rest were the default values. The conserved motifs and the gene structure of NtTALE genes were visualized by TBtools.
The cis-acting elements of the NtTALE promoters. The upstream 2000bp sequences of all NtTALE genes coding regions were extracted. The cis-acting elements were predicted by PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 2 April 2023) (Lescot et al. 2002). The result was visualized by the ggplot2 package.
Secondary and 3D structure of the NtTALE proteins. NtTALEs were predicted for protein secondary and 3D structures using the online websites SOMPA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa%20_sopma.html, accessed on 6 April 2023) and SWISS-MODEL (https://swissmodel.expasy.org/, accessed on 7 April 2023) (Waterhouse et al. 2018).
Expression patterns of the NtTALE genes. RNA-seq data were download from the SRA database (https://www.ncbi.nlm.nih.gov/sra, accessed on 23 April 2023) to analyze the expression patterns of NtTALE genes under different conditions, including different tissues (SRP029183), cold (SRP097876), dehydration (SRP301492), NaHCO3 (SRP193166), NaCl (SRP193166) and R. solanacearum (SRP336664).
SRA files were converted to FASTA files by Fastq-dump. Trimmomatic was used to remove adapters from the FASTQ sequence of the Illumina platform and to trim the FASTQ sequence file based on base mass values (Bolger et al. 2014). Hista2 was used to index and compare replies. SAM files were converted into BAM files by Samtools (Kim et al. 2019). The FPKM value was calculated by Stringtie and the count was calculated by the prepDE.py3 scriptt (Li et al. 2022). The DESeq2 package was used for differential analysis, and the differential expression gene screening criteria were: log2FoldChange ≥ 1 and padj ≤ 0.05 is up-regulated, log2FoldChange ≤ − 1 and padj ≤ 0.05 is down-regulated (Love et al. 2014). The results were visualized by TBtools. The Upset plot was made by the ChiPlot online website (https://www.chiplot.online/, accessed on 2 August 2023).
PPI of the NtTALE genes. The PPI of the NtTALE proteins were analyzed by the online website String (https://string-db.org/, accessed on 20 April 2023) (Szklarczyk et al. 2023). The network was beautified by Cytoscape software (Shannon et al. 2003).
Results
Identification and fundamental characteristics of the NtTALE genes. A total of 45 members of the NtTALE gene family were identified, including 18 members of the BELL subfamily and 27 members of the KNOX subfamily. They were renamed NtTALE1-45. The fundamental characteristics of the NtTALE genes were analyzed (Supplementary Table S1). The number of amino acids (AA) was 173 ~ 771, and the molecular weight (MW) of protein was 19509.86 ~ 84061.97 Da. The theoretical pI (pI) was 4.85 ~ 9.34, of which 39 NtTALE gene family members had a pI less than 7, which belongs to acidic proteins. The grand average of hydropathicity (GRAVY) was − 0.908 to − 0.407, all of which were negative, indicating that all proteins were hydrophilic proteins. The aliphatic index (AI) was 57.87 ~ 82.64.
Subcellular localization predictions found that all 45 NtTALE genes were localized to the nucleus. In addition, no signal peptides were detected in all any NtTALE genes. We speculated that this family member in tobacco might not have a transmembrane transport function. In summary, it was speculated that 45 NtTALE family proteins might all perform their biological functions in the nucleus.
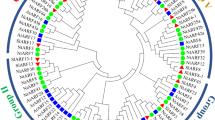
Phylogenetic and orthologous gene analysis of TALE genes. In order to analyze the classification of NtTALE genes and their phylogenetic relationships in Solanaceae, a phylogenetic tree was constructed using genes from 405 TALE genes in A. thaliana, rice and 11 Solanaceae species (Fig. 1). As a result, the phylogenetic tree was divided into two subfamilies, namely the BELL subfamily and the KNOX subfamily. The BELL subfamily has 27 NtTALE genes. The KNOX subfamily was further divided into KNOXI, KNOXII, KNOXIII, with ten, six, and two NtTALE genes, respectively. It is worth noting that tobacco has two KNOXIII subfamily genes (NtTALE29 and NtTALE45) that are not found in other Solanaceae plants. OrthoFinder analysis found that it could be divided into 13 orthologous gene groups (Supplementary Table 2, Supplementary Table 3). One of the orthologous groups is unique to regular tobacco and includes NtTALE29 and NtTALE45. Compared with Nicotiana tomentosiformis and Nicotiana sylvestris, some genes of Nicotiana tabacum were expanded and some genes were contracted (Supplementary Table 4).
Phylogenetic tree of the TALE genes. Nt, Nicotiana tabacum. Ns, Nicotiana sylvestris. Nto, Nicotiana tomentosiformis. At, Arabidopsis thaliana. Sl, Solanum lycopersicum. Ca, Capsicum annuum. Nb, Nicotiana benthamiana. Osj, Oryza sativa subsp. Japonica. Osi, Oryza sativa subsp. Indica. Sm, Solanum melongena. Spi, Solanum pimpinellifolium. Spt, Solanum ptychanthum. Spe, Solanum pennellii. St, Solanum tuberosum
Motifs compositions and gene structures of NtTALE genes. Motif analysis of 45 NtTALE genes using the online website MEME resulted in a total of 10 significant motifs (Fig. 2B; Supplementary Fig. S1). In the KNOX subfamily, KNOXI members all had motif 1, motif 3, except NtTALE30, other members had motif 6, motif 8. KNOXII members had motif 1, motif 3, motif 6, motif 10. Among them, NtTALE15 and NtTALE19 also had motif 7. NtTALE14, NtTALE17, NtTALE20, NtTALE37 also had motif 8. KNOXIII members had only motif 6 and motif 8. The BELL subfamily all had motif 2 and motif 4. Motif 1 was present in most NtTALE family members, and it might play a role in maintaining the basic functions of family members.
Motif composition and gene structure of NtTALE genes. A Phylogenetic tree of the NtTALE genes. B Conserved motifs of the NtTALE proteins. The colored squares represent different motifs. C Gene structures of the NtTALE genes. The black lines represent introns. UTR, untranslated regions; CDS, coding sequences. The scale bar at the bottom was used to estimate protein structure and gene structure size
The NtTALE gene family had a complex structure (Fig. 2C). The number of exons of in the NtTALE gene family was three to eight. In addition, except for NtTALE2, NtTALE22, NtTALE30, NtTALE33 did not have 5' UTR, other family members had 5' and 3' UTR, and some family members had multiple 5' UTRs. Differences in gene structure between NtTALE may lead to differences in their function.
The cis-acting elements of the NtTALE promoters. PlantCARE was uesd for cis-acting element prediction. After deleting the core promoters such as TATA-box and the unannotated promoters, the remaining elements were divided into three categories: light-responsive elements (Fig. 3A), elements that responsed hormonal and abiotic stresses (Fig. 3B), and plant development elements (Fig. 3C). There are a total of 53 elements, including 27 elements that respond to light, 11 elements that respond to hormones, four elements that respond to abiotic stress, and 15 elements that regulate plant development.
Among the hormone response elements, there were four kinds of auxin response elements, three kinds of gibberellin response elements, two kinds of methyl jasmonate response elements, one kinds of salicylic acid response element and one kinds of abscisic acid response element. There were also abiotic stress response elements that respond to low temperature, drought, wound, and defense and stress. And elements that could regulate plant development, including differentiation of the palisade mesophyll cell, meristem development, flavonoid biosynthesis, circadian rhythm control, etc. In summary, the NtTALE genes might be involved in hormonal regulatory responses and resistance to abiotic stresses.
Secondary and 3D structure of NtTALE proteins. The biological function of plant proteins is determined by the higher-level structure of proteins. NtTALE protein secondary structure prediction (Supplementary Fig. S2) revealed that the members of this family had alpha helix, beta turn, extended strand, random coil, mainly alpha helix and random coil, and beta turn and extended strand interspersed. Multiple alpha helixes were alternately arranged with beta turn to form a helix-turn-helix structure.
3D structure prediction was performed for all NtTALE proteins according to the template recommended by the website, retaining sequences with more than 30% similarity to the template, a total of 42. The 3D structure of proteins of the same subfamily are highly similar. For the other 3 proteins, namely NtTALE29, NtTALE30 and NtTALE45 had low confidence in their structural predictions, so the prediction results were not adopted in this study (Supplementary Fig. S3). It was observed that although there were differences in the structure of different NtTALE proteins, they all had spatial structures such as α-helix, β-turn, and random coil, and all had a helix-turn-helix structure. This is consistent with the helix-turn-helix structure of the HB gene.
Expression patterns of NtTALE genes in different tissues. The function of genes can be reflected through gene expression. According to the heatmap after downloading RNA-seq data analysis, the clustering results showed that the results could be divided into two branches. The analysis found that the NtTALE genes were expressed in each tissue, but there were obvious differences in expression in different tissues (Fig. 4). The most highly expressed genes were in the stem, and 11 NtTALE genes such as NtTALE16, NtTALE30 and NtTALE41 were highly expressed in the stem. Of the 11 NtTALE genes that were highly expressed in stems, seven belonged to the KNOXI subfamily and four belonged to the BELL subfamily. The least genes were expressed in dry capsule, with only NtTALE43 and NtTALE44 being more expressed. There was little difference in NtTALE genes expressed in different stages of leaves. However, some gene expression was expressed significantly at different stages of flowering. From im-mature flower to mature flower, the expression of genes such as NtTALE17 and NtTALE26 were upregulated, and the expression of genes such as NtTALE3 and NtTALE45 were down-regulated.
Expression patterns of NtTALE genes under abiotic and biotic stresses. Under different treatments, different NtTALE genes had different expression patterns. Under alkali stress (Fig. 5A), the expression of NtTALE6, NtTALE21, NtTALE25 and NtTALE37 were significantly regulated, and the expression of NtTALE10, NtTALE12, NtTALE27, NtTALE33, NtTALE36 and NtTALE38 were significantly down-regulated. After dehydration treatment (Fig. 5B), the expression of NtTALE3, NtTALE5, NtTALE6, NtTALE17, NtTALE25, NtTALE32, NtTALE37, NtTALE39, and NtTALE42 were significantly upregulated, and the expressions of NtTALE1, NtTALE4, NtTALE7, NtTALE9, NtTALE22, NtTALE23, NtTALE27, NtTALE33, NtTALE35, and NtTALE38 were significantly down-regulated. Under cold stress (Fig. 5C), the expressions of NtTALE5, NtTALE6, NtTALE17, NtTALE25, NtTALE27, NtTALE37, NtTALE38, and NtTALE42 were significantly upregulated, and the expressions of NtTALE3, NtTALE9, NtTALE15, NtTALE32, NtTALE36, and NtTALE40 were significantly down-regulated. Under salt stress (Fig. 5D), NtTALE37 expression was significantly upregulated and NtTALE27 expression was significantly down-regulated.
After Ralstonia solanacearum (R. solanacearum) infestation with tobacco (Fig. 5E). The expression of NtTALE37, NtTALE43 and NtTALE44 were significantly upregulated. The expression of NtTALE2, NtTALE4, NtTALE27 and other genes were significantly down-regulated.
PPI of the NtTALE genes. PPI are composed of proteins interacting with each other to participate in all aspects of life processes such as biological signaling, gene expression regulation, energy and material metabolism. In this study, the online website String was used to analyze of the NtTALE proteins (Fig. 6). Proteins that have interactions with other NtTALE members were preserved. Among them, NtTALE45 was the core members of the protein interaction of this family, and interact with multiple other members. NtTALE2, NtTALE6, NtTALE7, NtTALE9, NtTALE21, NtTALE29, and NtTALE41 are also key node proteins with important roles. In addition, the results of Gene Ontology (GO) enrichment provided by the String website showed that the family members were involved in multiple processes such as transcriptional regulation, meristem maintenance, and shoot system morphogenesis (Supplementary Table 5).
Discussion
The TALE gene family is a class of regulators composed of KNOX subfamily and BELL subfamily, which have an important regulatory role in plant growth and development. In this study, the cis-acting element, expression patterns, and phylogeny of NtTALE genes were comprehensively analyzed. A total of 45 NtTALE genes were identified in tobacco. From the perspective of physicochemical properties, there were great differences in AA and MW of 45 NtTALE genes, and the proteins were all hydrophilic proteins. Subcellular localization showed that all members of this family were located in the nucleus, which was consistent with the signal peptide prediction (Supplementary Table S1). This is consistent with the fact that the orthologous gene GmBLH4 of NtTALE33 is located within the nucleus (Supplementary Table S6) (Tao et al. 2018). The cis-acting elements showed that there were gibberellin, meristem development and other response elements in the promoter region of NtTALE genes (Fig. 3). We speculate that the NtTALE genes may be involved in the regulation of multiple hormone responses and plant growth and development. In previous research, Pisum sativum PsBELL1-2 could interact with the PsDELLA1 protein regulator of the gibberellin pathway, which previously played an important role in symbiotic development (Dolgikh et al. 2020). Poplar PagKNAT2/6b could directly inhibit the synthesis of gibberellin, altering plant architecture conditions, and improve drought resistance (Song et al. 2021). We observed that the promoter regions of the NtTALE1, NtTALE3, NtTALE12, NtTALE23, NtTALE32, NtTALE35, and NtTALE38 genes had gibberellin-responsive elements. In particular, the NtTALE32 gene of the BELL subfamily had multiple gibberellin-responsive elements. We hypothesize that these genes play an important role in gibberellin signaling.
The expression pattern of NtTALE gene differed under different treatments (Fig. 5 and Supplementary Fig. S4). In this study, we analyzed RNA-seq data from five stresses: alkali, dehydration, cold, salt, and R. solanacearum, with 10, 19, 14, two, and 17 genes significantly different from normal treatments, respectively. Among the five stresses, the most NtTALE genes were involved in response to dehydration treatment and the least number of genes responded to salt stress. It was found that multiple genes such as NtTALE6 and NtTALE38 had different roles in different stresses. It is worth noting that NtTALE27 and NtTALE37 showed different degrees of up-regulation and down-regulation of these five stresses, and it is speculated that NtTALE27 and NtTALE37 were core genes of the NtTALE genes, responding to multiple stresses of tobacco. Notably, NtTALE27 was only highly expressed in immature flowers (Fig. 4). NtTALE27 and ATH1 were homologous genes. Previous research has shown that ATH1 controls floral competency as a specific activator of FLOWERING LOCUS C expression (Proveniers et al. 2007). It also modulates growth at the interface between the stem, meristem, and organ primordia and contributes to the compressed vegetative habit of A. thaliana (Gomez-Mena and Sablowski 2008). We speculated that NtTALE27 was not only involved in the response to stress, but also has an important regulatory role in flower organs. The gene function of NtTALE27 could be further studied by gene overexpression or gene silencing (Koeppe et al. 2023; Erdoğan et al. 2023). Under salt stress(Fig. 5D), one gene expression was upregulated and one gene expression was down-regulated, which was very different from the number of TALE genes (8/18) identified in sweet orange (Peng et al. 2022). In addition, in wheat, TaKNOX11-A enhanced the drought and salt tolerance of A. thaliana, and TaKNOX11-A overexpressed plants reduced malondialdehyde content and increased proline content, enabling plants to adapt to unfavorable environments more effectively (Han et al. 2022). The function of NtTALE37 gene in salt stress can be verified by Crispr/cas9 gene editing technology in the future. It is worth noting that most of the genes that are significantly upregulated under abiotic stresses belong to the BELL subfamily. It is possible that members of the BELL subfamily play an important role in responding to abiotic stresses. Phylogenetic trees were constructed using TALE genes from A. thaliana, rice, and Solanaceae (Fig. 1). The NtTALE genes could be divided into four categories, namely BELL (27), KNOXI (10), KNOXII (6), KNOXIII (2) (Fig. 2A). In Phyllostachys edulis, the BELL subfamily had 31 members and the KNOX subfamily had 24 members (Que et al. 2022). Similar to the results of this study, there were significantly more members of the BELL subfamily than the KNOX subfamily. In addition, research has shown that heterodimeric complexes can form between members of two subfamilies to function (Fig. 6). In A. thaliana, BLH2/SAW1 and BLH4/SAW2 establish leaf shape by repressing growth in specific subdomains of the leaf at least in part by repressing expression of one or more of the KNOX genes (Bellaoui et al. 2001; Kumar et al. 2007; Hackbusch et al. 2005). Soybean GmBLH4 might heterodimerize with GmSBH1 to form functional complexes and function in modulating plant growth and development as well as in response to high temperature and humidity stress (Tao et al. 2018). We can study the interaction of members of the two subfamilies through yeast two-hybrid system to further understand the function of the NtTALE genes (Wang et al. 2022). The number of genes in Nicotiana tabacum increased by 2–29 compared to other species. OrthoFinder performs orthologous gene analysis. Nicotiana tabacum is compared with nightshade and capsicum species. Taking NtTALE2 and NtTALE41 as examples, it was found that there was only one orthologous gene in other species. The expansion of the NtTALE genes may be related to polyploidization. Research has shown that Nicotiana tabacum originated from Nicotiana sylvestris and Nicotiana tomentosiformis (Wang et al. 2024). Compared with Nicotiana sylvestris and Nicotiana tomentosiformis, some genes of Nicotiana tabacum were expanded and some genes were contracted (Supplementary Table 4). We speculated that Nicotiana tabacum has lost some genes during the long evolutionary process. The emergence of the KNOXIII subfamily (NtTALE29, NtTALE45) may be due to the fact that the protogene has the advantage of adapting to the environment and thus begins to evolve, the gene base site mutation, or other reasons (Carvunis et al. 2012). The KNOX-like unique gene KNATM found in A. thaliana was expressed in the proximal lateral domain of organ primordium and the boundary of mature organs (Magnani and Hake 2008). In tobacco, KNOXIII gene was only highly expressed in immature flowers (Fig. 4). NtTALE45 is also a key node protein of PPI (Fig. 6). We can further study the function of the NtTALE29 and NtTALE45 genes using genetic transformation and other means (Rahman et al. 2023).
Conclusion
The TALE genes are composed of KNOX subfamily and BELL subfamily, which play an important role in regulating the growth and development of plants. This study identified 45 tobacco TALE genes in tobacco. This study analyzed the expression patterns of NtTALE gene under different stresses, provided evidence of NtTALE gene expansion. This study further improved the research system of tobacco gene family and provided reference for the study of gene families of other species. It is helpful to explore excellent genetic resources and reveal the stress response mechanism. This study has important theoretical value and practical significance.
References
Ali H et al (2019) Genome wide identification and expression profiles of TALE genes in pineapple (Ananas comosus L). Tropical Plant Biol 12:304–317. https://doi.org/10.1007/s12042-019-09232-4
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:W39–W49. https://doi.org/10.1093/nar/gkv416
Bally J et al (2018) The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annu Rev Phytopathol 56:405–426. https://doi.org/10.1146/annurev-phyto-080417-050141
Bellaoui M et al (2001) The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13:2455–2470. https://doi.org/10.1105/tpc.010161
Belles-Boix E et al (2006) KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18:1900–1907. https://doi.org/10.1105/tpc.106.041988
Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG (1995) A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem 270:31178–31188. https://doi.org/10.1074/jbc.270.52.31178
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Brambilla V et al (2007) Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 19:2544–2556. https://doi.org/10.1105/tpc.107.051797
Brian, L. et al. A gene expression atlas for kiwifruit (Actinidia chinensis) and network analysis of transcription factors. BMC Plant Biol. 21. https://doi.org/10.1186/s12870-021-02894-x (2021).
Carvunis A-R et al (2012) Proto-genes and de novo gene birth. Nature 487:370–374. https://doi.org/10.1038/nature11184
Chen C et al (2023) TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant 16:1733–1742. https://doi.org/10.1016/j.molp.2023.09.010
Chen, G. et al. Characterization of Dof transcription factors and the heat-tolerant function of PeDof-11 in passion fruit (Passiflora edulis). Int. J. Mol. Sci. 24. https://doi.org/10.3390/ijms241512091 (2023).
Costa F et al (2020) KNOX protein KNAT1 regulates fruitlet abscission in litchi by repressing ethylene biosynthetic genes. J Exp Bot 71:4069–4082. https://doi.org/10.1093/jxb/eraa162
Desplan C, Theis J, O’Farrell PH (1988) The sequence specificity of homeodomain-DNA interaction. Cell 54:1081–1090. https://doi.org/10.1016/0092-8674(88)90123-7
Dolgikh AV, Rudaya ES, Dolgikh EA (2020) Identification of BELL Transcription factors involved in nodule initiation and development in the legumes Pisum sativum and Medicago truncatula. Plants 9. https://doi.org/10.3390/plants9121808 (2020).
Duvaud S et al (2021) Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res 49:W216–W227. https://doi.org/10.1093/nar/gkab225
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20. https://doi.org/10.1186/s13059-019-1832-y (2019).
Erdoğan İ, Cevher-Keskin B, Bilir Ö, Hong Y, Tör M (2023) Recent developments in CRISPR/Cas9 genome-editing technology related to plant disease resistance and abiotic stress tolerance. Biology 12. https://doi.org/10.3390/biology12071037.
Fernandez-Pozo N et al (2015) The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res 43:D1036–D1041. https://doi.org/10.1093/nar/gku1195
Gomez-Mena C, Sablowski R (2008) Arabidopsis thaliana homeobox gene1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 20:2059–2072. https://doi.org/10.1105/tpc.108.059188
Goodstein DM et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186. https://doi.org/10.1093/nar/gkr944
Hackbusch J, Richter K, Müller J, Salamini F, Uhrig JF (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci 102:4908–4912. https://doi.org/10.1073/pnas.0501181102
Hamant O, Pautot V (2010) Plant development: a TALE story. C R Biol 333:371–381. https://doi.org/10.1016/j.crvi.2010.01.015
Han Y et al (2022) Genome-wide analysis of TALE superfamily in Triticum aestivum reveals TaKNOX11-A is involved in abiotic stress response. BMC Genomics 23:89. https://doi.org/10.1186/s12864-022-08324-y
Hay A, Tsiantis M (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137:3153–3165. https://doi.org/10.1242/dev.030049
Horton P et al (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585-587. https://doi.org/10.1093/nar/gkm259
Hou J et al (2021) Genome-Wide analysis of the homeobox gene family and identification of drought-responsive members in Populus trichocarpa. Plants 10. https://doi.org/10.3390/plants10112284.
Kim D et al (2013) BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J 75:755–766. https://doi.org/10.1111/tpj.12236
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. https://doi.org/10.1038/s41587-019-0201-4
Koeppe S, Kawchuk L, Kalischuk M (2023) RNA interference past and future applications in plants. Int J Mol Sci 24. https://doi.org/10.3390/ijms24119755.
Kondhare KR, Vetal PV, Kalsi HS, Banerjee AK (2019) BEL1-like protein (StBEL5) regulates CYCLING DOF FACTOR1 (StCDF1) through tandem TGAC core motifs in potato. J Plant Physiol 241. https://doi.org/10.1016/j.jplph.2019.153014.
Kumar R et al (2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX Expression spatially in leaf margins. Plant Cell 19:2719–2735. https://doi.org/10.1105/tpc.106.048769
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054.
Lamesch P et al (2012) The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40:D1202-1210. https://doi.org/10.1093/nar/gkr1090
Lescot M et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Letunic I, Bork P (2021) Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. https://doi.org/10.1093/nar/gkab301
Li E et al (2012) The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 194:102–115. https://doi.org/10.1111/j.1469-8137.2011.04016.x
Li J, Shumate A, Wong B, Pertea G, Pertea M (2022) Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLOS Comput Biol 18. https://doi.org/10.1371/journal.pcbi.1009730 (2022).
Li S et al (2022) Genome-wide identification of wheat KNOX gene family and functional characterization of TaKNOX14-D in plants. Int J Mol Sci 23. https://doi.org/10.3390/ijms232415918 (2022).
Li D et al (2023) Functional study of Amorpha fruticosa WRKY20 gene in response to drought stress. Int J Mol Sci 24. https://doi.org/10.3390/ijms241512231.
Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379:66–69. https://doi.org/10.1038/379066a0
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15. https://doi.org/10.1186/s13059-014-0550-8.
Lu S et al (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. https://doi.org/10.1093/nar/gkz991
Ma Q et al. Genome-wide identification and characterization of TALE superfamily genes in cotton reveals their functions in regulating secondary cell wall biosynthesis. BMC Plant Biol 19. https://doi.org/10.1186/s12870-019-2026-1.
Magnani E, Hake S (2008) KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20:875–887. https://doi.org/10.1105/tpc.108.058495
Meng L et al (2018) BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. Plant J 94:1126–1140. https://doi.org/10.1111/tpj.13924
Nielsen H, Brunak S, von Heijne G (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng 12:3–9. https://doi.org/10.1093/protein/12.1.3
Niu X, Fu D (2022) The roles of BLH transcription factors in plant development and environmental response. Int J Mol Sci 23. https://doi.org/10.3390/ijms23073731.
Peng W et al (2022) TALE Transcription factors in sweet orange (Citrus sinensis): genome-wide identification, characterization, and expression in response to biotic and abiotic stresses. Front Plant Sci 12. https://doi.org/10.3389/fpls.2021.814252.
Proveniers M, Rutjens B, Brand M, Smeekens S (2007) The arabidopsis TALE homeobox geneATH1controls floral competency through positive regulation ofFLC. Plant J 52:899–913. https://doi.org/10.1111/j.1365-313X.2007.03285.x
Que F, Liu Q, Zha R, Xiong A, Wei Q (2022) Genome-wide identification, expansion, and evolution analysis of homeobox gene family reveals TALE genes important for secondary cell wall biosynthesis in moso bamboo (Phyllostachys edulis). Int J Mol Sci . https://doi.org/10.3390/ijms23084112.
Rahman SU, Khan MO, Ullah R, Ahmad F, Raza G (2023) Agrobacterium-mediated transformation for the development of transgenic crops; present and future prospects. Mol Biotechnol. https://doi.org/10.1007/s12033-023-00826-8
Sayers EW et al (2022) Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. https://doi.org/10.1093/nar/gkac1032
Shahan R, Li D, Liu Z (2019) Identification of genes preferentially expressed in wild strawberry receptacle fruit and demonstration of their promoter activities. Horticult Res 6. https://doi.org/10.1038/s41438-019-0134-6.
Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9:484–489. https://doi.org/10.1016/j.pbi.2006.07.008
Shannon P et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Sierro N et al (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5. https://doi.org/10.1038/ncomms4833.
Song X, Zhao Y, Wang J, Lu M-Z, Napier R (2021) The transcription factor KNAT2/6b mediates changes in plant architecture in response to drought via down-regulating GA20ox1 in Populus alba × P. glandulosa. J Exp Bot 72:5625–5637. https://doi.org/10.1093/jxb/erab201.
Szklarczyk D et al (2023) The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 51:D638–D646. https://doi.org/10.1093/nar/gkac1000
Tao Y et al (2018) Identification and functional characterization of a novel BEL1-LIKE homeobox transcription factor GmBLH4 in soybean. Plant Cell Tissue Organ Culture (PCTOC) 134:331–344. https://doi.org/10.1007/s11240-018-1419-4.
Tian F, Yang D-C, Meng Y-Q, Jin J, Gao G (2019) PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz1020
Wang J et al (2024) High-quality assembled and annotated genomes of Nicotiana tabacum and Nicotiana benthamiana reveal chromosome evolution and changes in defense arsenals. Mol Plant. https://doi.org/10.1016/j.molp.2024.01.008
Wang Y et al (2020) Genome-wide identification and expression analysis of TALE gene family in pomegranate (Punica granatum L.). Agronomy 10. https://doi.org/10.3390/agronomy10060829.
Wang S et al (2022) Protein‐protein interaction networks as miners of biological discovery. Proteomics 22. https://doi.org/10.1002/pmic.202100190.
Waterhouse A et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Xu P, Cai W (2019) Function of Brassica napus BnABI3 in Arabidopsis gs1, an Allele of AtABI3, in seed development and stress response. Front Plant Sci 10. https://doi.org/10.3389/fpls.2019.00067 (2019).
Xu X et al (2019) Identification of homeobox genes associated with lignification and their expression patterns in bamboo shoots. Biomolecules 9. https://doi.org/10.3390/biom9120862.
Yang Q, Yuan C, Cong T, Wang J, Zhang Q (2022) Genome-wide identification of three-amino-acid-loop-extension gene family and their expression profile under hormone and abiotic stress treatments during stem development of Prunus mume. Front Plant Sci 13:1006360. https://doi.org/10.3389/fpls.2022.1006360
Zhao K et al (2019) Comprehensive analysis of the three-amino-acid-loop-extension gene family and its tissue-differential expression in response to salt stress in poplar. Plant Physiol Biochem 136:1–12. https://doi.org/10.1016/j.plaphy.2019.01.003
Zhao W et al (2023) Alfalfa MsATG13 Confers cold stress tolerance to plants by promoting autophagy. Int J Mol Sci 24. https://doi.org/10.3390/ijms241512033.
Acknowledgements
This work was supported by the Foundation of Innovation Team Project for Modern Agricultural Industrious Technology System of Shandong Province (SDAIT-25-01) and the Foundation of Taishan brand cigarette high-quality core raw material development and application in Shandong (202102004). And we thank Supercomputing Center in Shandong Agricultural University for technical support.
Author information
Authors and Affiliations
Contributions
Conceived and designed the analysis: Y.S., K.S.; Performed the analysis: Y. S.; Drafting the manuscript: Y. S.; Critical revision of the manuscript: All authors. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, Y., Kong, D., Dai, Y. et al. Expression patterns and evidence of the expansion of Nicotiana tabacum L. TALE genes in the Solanaceae family. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02005-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02005-3