Abstract
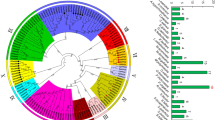
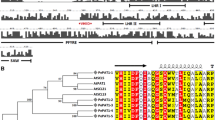
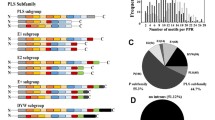
Populus trichocarpa (black cottonwood) is a fast-growing deciduous tree from the willow family (Salicaceae) grown for its fiber products and timber. Phototropism plays a substantial role in the growth, development, and survival strategies of poplar during stress responses. NPH3 (NONPHOTOTROPIC HYPOCOTYL 3) and RPT2 (ROOT PHOTOTROPISM 2), two members of the plant-specific NRL (NPH3/RPT2-like) gene family, are involved in phototropism. Although the NRL gene family has been characterized in most plant species with varying gene family sizes, the whole genome analysis of the NRL gene family in poplar has not been reported. In this study, 78 NRLs were identified, and their conserved domains and gene structures were meticulously characterized in poplar. Many cis-elements associated with plant growth and development, phytohormones, and stress receptiveness were identified in the promoters of PtNRLs. Phylogenetic and interaction network analyses indicated that the role of PtNRLs is evolutionarily conserved in Arabidopsis, rice, and poplar. To summarize, this research presents a detailed clarification of PtNRLs. It forms a basis for the functional analysis of PtNRL gene function and provides a reference for PtNRL gene family data.








Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Allu PR, Badrinarayanan L, Elchuri SV (2023) Light as an environmental factor for the well-being of the plant, animal, and human triad. One Health Hum an Environ Triad. https://doi.org/10.1002/9781119867333.ch29
An G, Qi Y, Zhang W, Gao H, Qian J, Larkin RM, Chen J, Kuang H (2022) LsNRL4 enhances photosynthesis and decreases leaf angles in lettuce. Plant Biotechnol J 20(10):1956–1967
Andolfo G, Sanseverino W, Rombauts S, Van de Peer Y, Bradeen J, Carputo D, Frusciante L, Ercolano MR (2013) Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytol 197(1):223–237
Ashfield T, Egan AN, Pfeil BE, Chen NW, Podicheti R, Ratnaparkhe MB, Ameline-Torregrosa C, Denny R, Cannon S, Doyle JJ (2012) Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol 159(1):336–354
Ban Z, Estelle M (2021) CUL3 E3 ligases in plant development and environmental response. Nat Plants 7(1):6–16
Bashir S, Rehman N, Fakhar Zaman F, Naeem MK, Jamal A, Tellier A, Ilyas M, Silva Arias GA, Khan MR (2022) Genome-wide characterization of the NLR gene family in tomato (Solanum lycopersicum) and their relatedness to disease resistance. Front Genet 13:931580
Brini F, Mseddi K, Brestic M, Landi M (2022) Hormone-mediated plant responses to light quality and quantity. Environ Exp Bot 202:105026
Chen W, Sheng Z, Cai Y, Li Q, Wei X, Xie L, Jiao G, Shao G, Tang S, Wang J (2019) Rice morphogenesis and chlorophyll accumulation is regulated by the protein encoded by NRL3 and its interaction with NAL9. Front Plant Sci 10:175
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Christie JM, Suetsugu N, Sullivan S, Wada M (2018) Shining light on the function of NPH3/RPT2-like proteins in phototropin signaling. Plant Physiol 176(2):1015–1024
de Carbonnel M, Davis P, Roelfsema MRG, Inoue S-I, Schepens I, Lariguet P, Geisler M, Shimazaki K-I, Hangarter R, Fankhauser C (2010) The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol 152(3):1391–1405
Dong C, Zhang L, Zhang Q, Yang Y, Li D, Xie Z, Cui G, Chen Y, Wu L, Li Z (2023) Tiller Number1 encodes an ankyrin repeat protein that controls tillering in bread wheat. Nat Commun 14(1):836
Dzinyela R, Alhassan AR, Suglo P, Movahedi A (2023) Advanced study of functional proteins involved in salt stress regulatory pathways in plants. S Afr J Bot 159:425–438
Gaba Y, Bhowal B, Pareek A, Singla-Pareek SL (2023) Genomic survey of flavin monooxygenases in wild and cultivated rice provides insight into evolution and functional diversities. Int J Mol Sci 24(4):4190
Haga K, Sakai T (2023) Photosensory adaptation mechanisms in hypocotyl phototropism: how plants recognize the direction of a light source. J Exp Bot 74(6):1758–1769
He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2012) Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep 31:1199–1217
Hu Y, Sun L, Wang D, Zhang D, Cai W (2022) Genome-wide identification and bioinformatics analysis of NRL gene family in rice. Mole Plant Breed. https://doi.org/10.5376/mpb.2022.13.0018
Huang Z, Qiao F, Yang B, Liu J, Liu Y, Wulff BB, Hu P, Lv Z, Zhang R, Chen P (2022) Genome-wide identification of the NLR gene family in Haynaldia villosa by SMRT-RenSeq. BMC Genomics 23(1):1–23
Hwarari D, Radani Y, Guan Y, Chen J, Liming Y (2023) Systematic characterization of GATA transcription factors in liriodendron chinense and functional validation in abiotic stresses. Plants 12(12):2349
Ishishita K, Higa T, Tanaka H, Inoue S-I, Chung A, Ushijima T, Matsushita T, Kinoshita T, Nakai M, Wada M (2020) Phototropin2 contributes to the chloroplast avoidance response at the chloroplast-plasma membrane interface. Plant Physiol 183(1):304–316
Jiang N, Yan J, Liang Y, Shi Y, He Z, Wu Y, Zeng Q, Liu X, Peng J (2020) Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.)–an updated review. Rice 13(1):1–12
Ke Y, Xu M, Hwarari D, Chen J, Yang L (2022) Genomic survey of heat shock proteins in liriodendron chinense provides insight into evolution, characterization, and functional diversities. Int J Mol Sci 23(23):15051
Kimura M, Kagawa T (2006) Phototropin and light-signaling in phototropism. Curr Opin Plant Biol 9(5):503–508
Legris M, Boccaccini A (2020) Stem phototropism toward blue and ultraviolet light. Physiol Plant 169(3):357–368
Li Y, Dai X, Cheng Y, Zhao Y (2011) NPY genes play an essential role in root gravitropic responses in Arabidopsis. Mol Plant 4(1):171–179
Li J, Su X, Wang Y, Yang W, Pan Y, Su C, Zhang X (2018) Genome-wide identification and expression analysis of the BTB domain-containing protein gene family in tomato. Genes Genom 40:1–15
Li M, Hwarari D, Li Y, Ahmad B, Min T, Zhang W, Wang J, Yang L (2022a) The bZIP transcription factors in Liriodendron chinense: Genome-wide recognition, characteristics and cold stress response. Front Plant Sci 13:1035627
Li R, Ahmad B, Hwarari D, Li DA, Lu Y, Gao M, Chen J, Yang L (2022b) Genomic survey and cold-induced expression patterns of bhlh transcription factors in liriodendron chinense (Hemsl) Sarg. Forests 13(4):518
Liang X, Dong J (2023) Comparative-genomic analysis reveals dynamic NLR gene loss and gain across Apiaceae species. Front Genet 14:1141194
Lidder P, Sonnino A (2012) Biotechnologies for the management of genetic resources for food and agriculture. Adv Genet 78:1–167
Lin X, Jia Y, Heal R, Prokchorchik M, Sindalovskaya M, Olave-Achury A, Makechemu M, Fairhead S, Noureen A, Heo J (2023) Solanum americanum genome-assisted discovery of immune receptors that detect potato late blight pathogen effectors. Nat Genet 55:1579–1588
Liscum E, Askinosie SK, Leuchtman DL, Morrow J, Willenburg KT, Coats DR (2014) Phototropism: growing towards an understanding of plant movement. Plant Cell 26(1):38–55
Liu L, Xu J, Chen J, Yang B, Yang C, Yang Y, Wang K, Zhuo R, Yao X (2023) The LEA2 gene sub-family: characterization, evolution, and potential functions in Camellia oleifera seed development and stress response. Sci Hortic 322:112392
Lopez Vazquez A, Allenbach Petrolati L, Legris M, Dessimoz C, Lampugnani ER, Glover N, Fankhauser C (2023) Protein S-acylation controls the subcellular localization and biological activity of PHYTOCHROME KINASE SUBSTRATE. Plant Cell 35(7):2635–2653
Manasra S, Kajava AV (2023) Why does the first protein repeat often become the only one? J Struct Biol 215(3):108014
Movahedi A, Dzinyela R, Aghaei-Dargiri S, Alhassan AR, Yang L, Xu C (2023) Advanced study of drought-responsive protein pathways in plants. Agronomy 13(3):849
Naqvi S, He Q, Trusch F, Qiu H, Pham J, Sun Q, Christie JM, Gilroy EM, Birch PR (2022) Blue-light receptor phototropin 1 suppresses immunity to promote Phytophthora infestans infection. New Phytol 233(5):2282–2293
Petersen J, Inoue S-I, Kelly SM, Sullivan S, Kinoshita T, Christie JM (2017) Functional characterization of a constitutively active kinase variant of Arabidopsis phototropin 1. J Biol Chem 292(33):13843–13852
Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genomics 281:609–626
Reuter L, Schmidt T, Manishankar P, Throm C, Keicher J, Bock A, Droste-Borel I, Oecking C (2021) Light-triggered and phosphorylation-dependent 14-3-3 association with NON-PHOTOTROPIC HYPOCOTYL 3 is required for hypocotyl phototropism. Nat Commun 12(1):6128
Salmirinne IO (2022) Bioinformatic and biochemical characterization of arabidopsis thaliana protein nch1 and links to plant immunity. University of Glasgow.
Seo E, Kim S, Yeom S-I, Choi D (2016) Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding leucine-rich repeat gene family among Solanaceae plants. Front Plant Sci 7:1205
Sonnino A (2015) Current status of biotechnology development and application in forestry, In: Proceedings of the international symposium on forest biotechnology for smallholders, Foz do Iguaçu, Paraná, Brazil. pp 19–22.
Sonnino A (2016) Current and potential application of biotechnology in forestry: a critical review. Asian Biotechnol Dev Rev 18(3).
Suetsugu N, Takemiya A, Kong S-G, Higa T, Komatsu A, Shimazaki K-I, Kohchi T, Wada M (2016) RPT2/NCH1 subfamily of NPH3-like proteins is essential for the chloroplast accumulation response in land plants. Proc Natl Acad Sci 113(37):10424–10429
Sullivan S, Waksman T, Paliogianni D, Henderson L, Lütkemeyer M, Suetsugu N, Christie JM (2021) Regulation of plant phototropic growth by NPH3/RPT2-like substrate phosphorylation and 14-3-3 binding. Nat Commun 12(1):6129
Sunarti S, Kissoudis C, Van Der Hoek Y, Van Der Schoot H, Visser RG, Van Der Linden CG, Van De Wiel C, Bai Y (2022) Drought stress interacts with powdery mildew infection in tomato. Front Plant Sci 13:845379
Taylor G (2002) Populus: arabidopsis for forestry. Do we need a model tree? Ann Bot 90(6):681–689
Tsutsumi T, Takemiya A, Harada A (2013) Disruption of ROOT PHOTOTROPISM2 gene does not affect phototropin-mediated stomatal opening. Plant Sci 201:93–97
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A (2006) The genome of black cottonwood Populus trichocarpa (Torr. & Gray). Science 313(5793):1596–1604
Waksman T, Suetsugu N, Hermanowicz P, Ronald J, Sullivan S, Łabuz J, Christie JM (2023) Phototropin phosphorylation of ROOT PHOTOTROPISM 2 and its role in mediating phototropism, leaf positioning, and chloroplast accumulation movement in Arabidopsis. Plant J 114(2):390–402
Wan H, Yuan W, Ye Q, Wang R, Ruan M, Li Z, Zhou G, Yao Z, Zhao J, Liu S (2012a) Analysis of TIR-and non-TIR-NBS-LRR disease resistance gene analogous in pepper: characterization, genetic variation, functional divergence and expression patterns. BMC Genom 13:1–15
Wan Y, Jasik J, Wang L, Hao H, Volkmann D, Menzel D, Mancuso S, Baluška F, Lin J (2012b) The signal transducer NPH3 integrates the phototropin1 photosensor with PIN2-based polar auxin transport in Arabidopsis root phototropism. Plant Cell 24(2):551–565
Wei H, Wang P, Chen J, Li C, Wang Y, Yuan Y, Fang J, Leng X (2020) Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol 20:1–19
Xin G-Y, Li L-P, Wang P-T, Li X-Y, Han Y-J, Zhao X (2022) The action of enhancing weak light capture via phototropic growth and chloroplast movement in plants. Stress Biol 2(1):50
Xu W, Jian S, Li J, Wang Y, Zhang M, Xia K (2023) Genomic identification of CCCH-type zinc finger protein genes reveals the role of HuTZF3 in tolerance of heat and salt stress of pitaya (Hylocereus polyrhizus). Int J Mol Sci 24(7):6359
Yang L, McLellan H, Naqvi S, He Q, Boevink PC, Armstrong M, Giuliani LM, Zhang W, Tian Z, Zhan J (2016) Potato NPH3/RPT2-like protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiol 171(1):645–657
Yang M, Wang L, Chen C, Guo X, Lin C, Huang W, Chen L (2021) Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress. Sci Rep 11(1):22933
Zhu Y, Wu N, Song W, Yin G, Qin Y, Yan Y, Hu Y (2014) Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol 14:1–19
Acknowledgements
We thank all the researchers who helped us gather references and those who reviewed the manuscript.
Funding
This study was supported by a start-up foundation at Nanjing Forestry University (No. 163108059).
Author information
Authors and Affiliations
Contributions
R.D. designed and drafted the manuscript and performed the analysis; D.H. assisted with the analysis and interpretation of the data; A.R.A. and P.S. revised the manuscript; A.M. reviewed, edited, funded, and supervised this project. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dzinyela, R., Hwarari, D., Alhassan, A.R. et al. Bioinformatics and expression analysis of the NRL gene family in Populus trichocarpa. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02003-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02003-5