Abstract
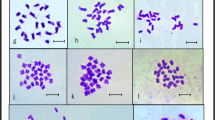
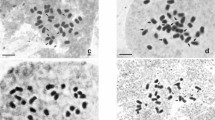
Indian Squill (Drimia indica) belonging to the family Asparagaceae is a highly medicinal herb with several therapeutic properties. The cytotaxonomic complexity of the species Drimia makes it intriguing to investigate ploidy variation in ecotype level and its significance in relation to genome size variation. Detailed karyotype analysis and 2C DNA contents were reported in twelve ecotypes of D. indica from different geographical locations of the state Odisha, India. The diploid somatic chromosome 2n = 2x = 20 was recorded in four ecotypes such as ‘Kendrapada’, ‘Tikarpada’, ‘Daspalla’, and ‘Odagaon, while tetraploid having 2n = 4x = 40 was newly reported in ecotype from ‘Nuagaon’. Aneuploid chromosome number 2n = 26 was obtained in ecotypes ‘Similipal’ and ‘Satkosia’ and 2n = 32 was newly reported in ecotypes ‘Bitarkanika’ and ‘Narasinghpur’. The ecotypes ‘Nayagarh’ and ‘Nilagiri’ were recorded with 2n = 16 and 2n = 22 respectively. The 2C DNA content analysis showed 2.19-fold increase which correlates with ploidy variation in ecotype level. The 2C DNA values were found highest of 36.26 pg (2n = 40) in ecotype ‘Nuagaon’ and lowest at 16.54 pg (2n = 16) in ecotype ‘Nayagarh’ that corresponds with ploidy and genome size. The asymmetry in the karyotypes showed non-significant clusters among the ecotypes except ecotypes ‘Odagaon’ and ‘Bhitarkanika’ which are most asymmetric having advanced karyotypic features as compared to the ecotypes ‘Simlipal’, ‘Karanjia’ and ‘Nayagarh’ having symmetric karyotypes. The chromosome characteristics along with genome size analysis of D. indica can be very useful in establishing genome-specific cytotypes which can be used for breeding programme for crop improvement. Furthermore, enhancement of therapeutic phyto-constituents, and cyto-taxonomic identification from adulterant and confirmation of alloploidy or autoploidy through genomic in situ hybridization could unravel the potential of this plants in relation to secondary metabolite production.






Similar content being viewed by others
References
Ansari MY, Raghavan RS (1980) Nomenclature of some bulbous Liliaceae of India. J Bombay Nat Hist Soc 77:172–173
Aswal S, Kumar A, Semwal RB, Chauhan A, Kumar A, Lehmann J, Semwal DK (2019) Drimia indica: a plant used in traditional medicine and its potential for clinical uses. Medicina 55:255–271
Baquar SR (1989) Medicinal and poisonous plants of Pakistan. Printas; Karachi, Pakistan
Bennett M, Leitch I (2005) Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot 95:45–90
Bennett MD, Johnston S, Hodnett GL, Price HJ (2000) Allium cepa L. cultivars from four continents compared by flow cytometry show nuclear DNA constancy. Ann Bot 85:351–357
Chittoor MS, Binny AJR, Yadlapalli SK, Cheruku A, Dandu C, Nimmanapalli Y (2012) Anthelmintic and antimicrobial studies of Drimia indica (Roxb.) Jessop. bulb aqueous extracts. J Pharm Res 5:3677–3686
Das AB, Das P (1997) Estimation of nuclear DNA and karyotype analysis in nine cultivars of Musa acuminate. Cytobios 90:181–192
Das AB, Mallick R (1993) Karyotype diversity and interspecific 4C DNA variation in Bupleurum. Biol Plant 35:355–363
Das AB, Dehery SK, Jena SN, Sinha RK (2020) A new seeded diploid accession of Musa laterita of section Rhodochlamys fom Gangtok, Sikkim, India with morphology, chromosome number and genome size. Cytologia 85:63–69
Dehery SK, Das AB (2022) Genetic diversity of twelve triploid bananas and plantains under section Eumusa as evident by chromosome morphology and SSR markers. Nucleus 65:35–48
Dehery SK, Panda E, Saha PR, Sinha RK, Das AB (2021) Chromosome diversity and karyotype asymmetry analysis in four cultivated triploid and three diploid wild genotypes of Musa from North-East India. Nucleus 64:167–179
Desai N, Kawalkar H, Dixit GB (2012) Biosystematics and evolutionary studies in Indian Drimia species. J Syst and Evol 50:512–518
Divya AJ, Venkataramegowda S (2021) Analysis of nuclear DNA content in plant cells of Urginea wightii by flow cytometry. Plant Cell Biotechnol Mol Biol 22:47–53
Doležel J, Doleželová M, Novák FJ (1994) Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol Plantarum 36:351–357
Doyle GG (1986) Aneuploidy and inbreeding depression in random mating and self-fertilizing autotetraploid populations. Theor Appl Genet 72:799–806
Geetha HL, Shivakameshwari MN, Nijagunaiah R (2016) Karyotypic variations in different accessions of Urginea indica (Roxb) Kunth. and Urginea wightii Lakshmin. Hyacinthaceae Cytologia 81:329–334
Guerra M (2008) Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenet Genome Res 120:339–350
Imai HT, Maruyama T, Gojobori T, Inoue Y, Crozier RH (1986) Theoretical bases for karyotype evolution. I. the minimum-interaction hypothesis. Am Nat 128:900–920
Lekhak MM, Adsul AA, Yadav S (2014) Cytotaxonomical studies on Drimia nagarjunae and a review on the taxonomy of Indian species of Drimia. Nucleus 57:99–103
Lekhak MM, Yadav PB, Yadav SR (2017) Cytogenetic studies in Indian Drimia Jacq. (Urgineoideae: Hyacinthaceae). In: Bhat TA, Wani AA (eds) Chromosome Structure and Aberrations. Springer, India, pp 141–165
Levan A, Fredya K, Sandberg A (1964) Nomenclatyure for centromeric position on chromosome. Heridity 52:201–220
Liang G, Chen H (2015) Scaling chromosomes for an evolutionary karyotype: a chromosomal trade off between size and number across woody species. PLoS ONE 10:e0144669
Nath S, Jha TB, Mallick SK, Jha S (2015) Karyological relationships in Indian species of Drimia based on fluorescent chromosome banding and nuclear DNA amount. Protoplasma 252:283–299
Pandey D, Gupta AK (2014) Antimicrobial activity and phytochemical analysis of Urginea indica from Bastar district of Chhattisgarh. Int J Pharm Sci Rev Res 26:273–281
Paszko B (2006) A critical review and a proposal of karyotype asymmetry indices. Plant Syst Evol 258:39–48
Patel P, Dehery SK, Pradhan C, Das AB (2019) Ecotype diversity of numerical and structural chromosomes of Drimia indica (Roxb.) Jessop (Hyacinthaceae) as revealed from karyotype analysis. Plant Sci Research 41:1–7
Peruzzi L, Eroǧlu HE (2013) Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet 7:1–9
Prajapathi ND, Purohit SS, Sharma AK, Kumar TA (2003) Handbook of medicinal plants: a complete source book. Books of Agrobios, India, p 396
Shiva Kameshvari MN (2004) Biosystematic studies and conservation of Urginea indica Kunth. Liliaceae J Swamy Bot Club 21:1–8
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold, London, p 216
Vimala Y, Lavania S, Lavania UC (2021) Chromosome change and karyotype differentiation–implications in speciation and plant systematics. Nucleus 64:33–54. https://doi.org/10.1007/s13237-020-00343-y
Yadav PB, Manning JC, Yadav SR, Lekhak MM (2019) A cytotaxonomic revision of Drimia Jacq. (Hyacinthaceae: Urgineoideae) in India. S Afr J Bot 123:62–86
Acknowledgements
We are grateful to Prof. A. Pahdi, Head, Department of English, Utkal University, for English correction of the manuscript. The internal standard materials received from Prof. J. Doležel, Institute of Experimental Botany, Olomouc, The Czech Republic used in flow cytometry is gratefully acknowledged. Administrative support of Utkal University and use of microscopic facilities developed under DSR-III, University Grant Commission, and FIST program, Govt. of India at Department of Botany is acknowledged to carry out the research. The use of the flow-cytometric facility in CSIR-National Botanical Research Institute, Lucknow is acknowledged. PP acknowledge the receipt of financial assistance from Biju Patnaik Research fellowship (No. ST-SCST-MISC-0054-2018/1152/ST dt. 26.02.2019), Science Technology Department, Govt. of Odisha. ABD acknowledges funding received under Emeritus Professorship from Human Resource Development Group, Council of Industrial Research (Scheme No. 21(1107)/20/EMR-II), Ministry of Science and Technology, Government of India.
Funding
Financial assistance received from Science and Technology Department, Govt. of Odisha and CSIR, Govt. of India.
Author information
Authors and Affiliations
Contributions
ABD—Experimental design and concept of paper, karyogram data analysis, 2C data graph preparation and final manuscript editing; PP, SKD—Chromosome preparation, data recording, calculation, first draft manuscript preparation; SRJ—Flow-cytometry data recording, analysis and manuscript corrections; CP—Statistical analysis and photograph arrangement and text corrections.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflict of any interest in this paper. All authors read the manuscript and approve the submission of this paper in this journal. The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Patel, P., Dehery, S.K., Jena, S.N. et al. Chromosomal variations in twelve ecotypes of a medicinal plant Drimia indica (Roxb.) Jessop: karyotypes and 2C DNA content analysis. Genet Resour Crop Evol 71, 621–634 (2024). https://doi.org/10.1007/s10722-023-01645-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01645-1