Abstract
1-deoxynojirimycin (DNJ) has certain medicinal value in lowering blood sugar, lowering blood fat, antiviral and anti-tumor, and Mulberry is one of known species with the highest DNJ content. To take full advantage of medicinal benefits of mulberry, the internal relationships of the genetic diversity of SSR markers and DNJ content was assessed in this study, so as to provide the basis for the exploitation and breeding of mulberry resources with high DNJ content. The results showed that the DNJ content in the leaves of the 36 mulberry germplasm resources varied significantly, with the mean value of 1.2794 mg/g, the amplitude of 1.03–1.61 g/g, and the coefficient of variation of 0.1276. Polymorphism was found in by 5 pairs of SSR primers from 36 mulberry germplasm resources, and a total of 32 genotypes were deteced. The mean allele number was 3.8, the mean heterozygosity was 0.62, the polymorphic information content of 0.61. Cluster analysis based on the genetic distance of SSR markers showed that the tested 36 mulberry germplasm resources could be divided into 3 class, and the analysis based on DNJ content showed that they could be divided into 2 clades. Comparison of the two cluster analyses displayed that the DNJ content in mulberry germplasm resources of the same group was similar. The above results revealed that the genetic diversity of SSR markers was similar to the genetic diversity of DNJ content in mulberry leaf.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction 1-Deoxynojirimycin (DNJ) is a class of piperidine alkaloid widely distributed in nature and revealed the inhibitory activities against α-glucosidase (Dong et al. 2018). Modern pharmacological experiments have also indicated that DNJ possesses an excellent regulatory effect to blood glucose, cholesterol, and triglycerides (Yan et al. 2015), promising a widespread applications in the treatment of diabetes and other diseases. Mulberry (Morus alba L.) was the known plant containing the highest content of DNJ, with the majority of DNJ residing in the leaves, branches, and roots (Zhou et al. 2018). Relevant data show that, its leaves are the main source of natural DNJ,but DNJ content of mulberry leaves is affected by many factors including the leaf position, picking period, growth condition, and germplasm resources, among other factors(Hao et al. 2018; Liu et al. 2006; Tao et al. 2018; Liu et al. 2012; Shi et al. 2013; Zhang et al. 2018a, b). It is stated that significant differences in DNJ content among mulberry varieties. And it is practically significant for application of DNJ to determining mulberry varieties with an above-average DNJ content.
According to incomplete statistics, there have been more than 1000 bred mulberry varieties and cultivated across 28 provinces in China. Moreover, the genetic diversity for agronomic, physiological, and nutritional traits of mulberry have evolved during the long-term selection and evolutionary process because of differences in planting environments, planting mode, fertilizer application, and water conditions (Peng et al. 2010). Genetic diversity of mulberry germplasm has been assessed using ISSR (inter simple sequence repeat), ALFP (amplified fragment length polymorphism), SSR (simple sequence repeat), and other molecular markers by numerous scholars globally. These studies postulate that the genetic relationship among mulberry germplasm resources is significantly related to geographical distribution, ecotype (Zhao et al. 2008), origin (Botton et al. 2005), and yield economy (Vijayan et al. 2006; Zhao et al. 2006). However, there was no reports of the relationship between the genetic diversity and the nutritional components of mulberry, such as the DNJ content. Aboved all, This study assessed the genetic diversity of mulberry varieties using SSR markers and further correlated genetic diversity and DNJ content to lay a foundation for subsequent breeding, development, and utilization of mulberry varieties with high DNJ content. The study provides invaluable information for strengthening the medicinal value and application of DNJ.
Materials and methods
Information on mulberry varieties
This study assessed 36 tested mulberry varieties planted in the garden of Jiangxi Sericulture and Tea Research Institute, at a latitude of 28°22′19″ N., longitude of 116°0′6″ W., and an altitude of 78 m.a.s.l. The information of the tested mulberry varieties is outlined in Table 1.
Collection of mulberry leaves
Mulberry leaves were collected on the morning of 2nd August 2019. The third leaf from the whole plant of each variety was sampled, and then 2000 g of fresh disease-free leaves of each variety were placed in a fresh-keeping bags (25 m × 35 m) and marked. Sub-samples of 50 g of leaves for each variety were stored at − 80 ℃ for genomic DNA extraction and subsequent SSR molecular marker analysis. The remaining mulberry leaves were dried at 60 ℃ for 72 h and then crushed into powder to determine the DNJ content.
Detection of DNJ content in mulberry leaves
Mulberry leaf powder (1 g) was dissolved in 20 mL hydrochloric acid (0.05 mol/L) in a 50 mL centrifuge tube, followed by extraction through vortexing for 1 min, ultrasonication for 20 min, and centrifugation at 12,000×g for 5 min. The extraction steps were repeated once, both supernatants were subsequently combined, and the volume topped up to 10 mL using 0.05 mg/L hydrochloric acid. The extracts were then subjected to HPLC after derivatization, following the methods described by Yu et al. (2019).
SSR marker analysis. Genomic DNA extraction and determination of quality
The genomic DNA of the mulberry varieties was extracted from its leaves using the Ezup column animal genomic DNA extraction kit (Shanghai Bioengineering Co., Ltd.). The DNA quality was subsequently checked on a 1% agarose gel using electrophoresis at 300 V for 30 min, and the DNA bands were visualized using a WFH-201B ultraviolet reflector. The DNA concentration was determined using a nanodrop ultraviolet spectrophotometer and stored at − 20 ℃.
Primer screening
Five pairs of core primers with a rich polymorphism, good repeatability, and clear bands were selected from ten pairs of primers reported by Peng et al. (2010). The primer information is outlined in Table 2. They were synthesized by Anhui general biology Co., Ltd. CTGTAAAACGGCCAGT was added to the 5′ end of all upstream primers for fluorescence (FAM) labelling.
PCR reaction and product detection
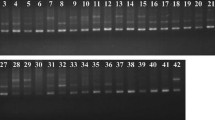
The PCR reaction mix contained 5 µL of 2 × Taq PCR Master Mix composed of dNTPs, Taq polymerase, and MgCl2 (Beijing Tiangen Co., Ltd), 2 µL DNA template (5–10 ng/µL), 0.1 µL 5′ M13-tailed forward primer (10 µM), 0.3 µL fluorescent labeling primer (FAM) (10 µM), 0.4 µL the reverse primer (10 µM), and 2.2 µL ultra-pure water totaling to 10 µL. The PCR conditions used were initial denaturation at 94 ℃ for 5 min, followed by 40 cycles of denaturation, annealing, and extension at 94 ℃ for 60 s, 50 ℃ for 40 s, and 72 ℃ for 90 s, respectively, and a final extension at 72 ℃ for 10 min. The PCR products were subsequently detected by capillary electrophoresis following the steps described by Guo et al. (2017).
Statistical analysis
Quantification of DNJ in the mulberry leaves was done using the external standard method, and the linear regression equation of the DNJ standard curve was: y = 3.253x + 2.076 (R2 = 0.9999), where X is the peak area and Y is the mass concentration of DNJ (µg/mL). All alleles of the SSR markers were regarded as the dominant markers and were coded as 1 for presence and 0 for absence. The genetic diversity parameters were calculated using the Power Marker v. 3.25 software following the methods described by Liu et al. (2005). The parameters included the number of observed alleles (NA), genotype number, gene diversity, heterozygosity, polymorphism information content (PIC), and Nei’s genetic distance. The parameters were subsequently used to construct a genetic distance matrix of the 36 varieties. Cluster analyses were carried out using the DPS software v.9.45 (wang et al. 2014). Systematic clustering of the SSRs among the 36 mulberry cultivars followed the Euclidean distance. It adopted the cluster average method in which the 36 genetic distance matrices were used as samples in the logarithmic transformation of the data. Similarly, cluster analysis of the DNJ content employed the cluster average method and adopted the 36 mulberry germplasm materials as indicators and their DNJ contents as samples to ensure data centralization. The Euclidean distance was used as the clustering scale.
Results and discussion
Evaluation of the DNJ content in mulberry leaves
The DNJ contents of the 36 mulberry cultivars (Table 3) were obtained by comparing the standard (Fig. 1) and the tested chromatograms (Fig. 2). There were significant differences in the DNJ content of the 36 mulberry germplasm resources, ranging between 1.03 µg/g and 1.61 µg/g. The mean value of the DNJ contents of the mulberry varieties was 1.2794 µg/g, with a coefficient of variation of 0.1276. Morus alba variety had the highest DNJ content at 1.61 µg/g, while Husang 199 had the lowest DNJ content at 1.03 µg/g. Eight mulberry varieties: zhiduosang, Husang 199, xiang7920, zhongsang 5801, dahuasang, xinyizhilai, and Jianchi had DNJ contents less than 1.10 µg/g. In addition, ninevarieties: Heye Husang, Husang 86, Husang 13, Sanghai 1, Dashi, Lunjiao 504, tiansang, and Nangang 2 had DNJ contents between 1.10 µg/g and 1.30 µg/g. The remaining 19 varieties had DNJ contents of more than 1.30 µg/g, with five and three varieties having more than 1.4 µg/g and 1.5 µg/g, respectively.
Previous studies postulate that DNJ is an important bioactive substance for evaluating the hypoglycemic activity in Mulberry (Morus L.) and silkworm (Bombyx mori). It is also an important index for measuring the medicinal value of mulberry and silkworm. Notably, DNJ is present in all mulberry leaves, but its content varies considerably among varieties, across growth periods, and in different cultivation conditions (Shi et al. 2013; Zhao et al. 2019; Zhang et al. 2018a, b; Zeng et al. 2018). These variations are potentially attributed to the genetic and physiological characteristics of the varieties, along with climatic and cultivation conditions, such as light, temperature, humidity, rainfall, and soil type, which influence the production of secondary metabolites in plants (Lou et al. 2011).
Genetic diversity analysis of mulberry leaves using SSR Markers
SSR marker analysis revealed 32 genotypes scored by the five selected primers pairs with an average of 6.4 bands per primer (Table 4). There were 19 alleles with an average of 3.80 alleles, a maximum of 5 alleles, and a minimum of 3 alleles per primer. The maximum and minimum heterozygosity of the primers was 1 (TP1) and 0.53 (TP2 and TP5), respectively, with an average of 0.62. The primers’ polymorphic information content (PIC) ranged between 0.52 and 0.66, with an average value of 0.61, displaying high polymorphism and an elaborate expression of the genotypic differences of the mulberry varieties. Moreover, the genetic diversity index of each primer pair ranged between 0.57 and 0.70, with an average of 0.61. These findings were consistent with those of Liu et al. (2019), which reported a gene diversity index ranging between 0.5548 and 0.6003 in 89 mulberry varieties using 14 SSR primer pairs. However, they were inconsistent with those of Peng et al. (2010), which reported a genetic diversity index ranging between 0 and 0.2015 in 172 mulberry cultivars using 10 SSR primer pairs. The differences are attributed to the number of primers used and the primers selected, which induced different diversities based on the different loci variations.
Evaluation of the genetic diversity of Mulberry germplasm resources
The genetic diversity of the tested mulberry germplasm resources was evaluated using the cluster analysis of the genetic distance of SSR markers and the DNJ content. The cluster analysis based on the the SSR markers showed that the tested 36 mulberry varieties could be divided into 3 class: 23 varieties in class I, 7 in class II, and 6 in class III (Fig. 3). In addition, the varieties were divided into two clades based on the DNJ content: clade I comprising 11 varieties and clade II comprising 25 cultivars (Fig. 4). The clades were further divided into four sub-clades: sub-clades I–VI. Notably, most of the varieties in class II in Fig. 3 were highly consistent with those in clade I in Fig. 4. These varieties had a low DNJ content ranging between 1.03 µg/g and 1.12 µg/g, with an average of 1.071 µg/g. Moreover, sub-clade I in Fig. 4 included six mulberry varieties with an average DNJ content of 1.258 µg/g, which was consistent with varieties in class III in Fig. 3. Sub-clades II and III had seven and nine varieties, with an average DNJ content of 1.336 µg/g and 1.58 µg/g, respectively. These results suggested similarities in the DNJ content of Mulberry varieties in the same class and vice versa, indicating some similarities between the DNJ content and polymorphism of the SSR molecular markers. These similarities were attributed to the strong genetic expression of DNJ regulatory genes in mulberry leaves during mulberry breeding. The lysine decarboxylase (MaLDC) and copper amine oxidase (MaCAO) are the key enzyme-coding genes involved in the biosynthesis of DNJ. The gene expression level and enzyme activity of the two enzymes are significantly positively correlated with the DNJ content in mulberry leaves (Wang et al. 2018). Transcriptome sequencing of two mulberry varieties with different DNJ contents suggests that lysine forms cadaverine through MaLDC, which is then oxidized to Δ1-piperideine under the action of MaCAO. DNJ is finally synthesized through a series of enzymatic reactions (Wang et al. 2018). Though this study suggests similarities in the genetic diversity of mulberry using SSR markers and based on the DNJ content, further exploration of the specific SSR markers and genes regulating the DNJ content should be done.
Cluster analysis of 36 tested mulberry germplasm resources based on the genetic distance of fluorescent SSR markers. (Note) germplasm resource No. are the same to those in Table 1
Cluster analysis of 36 tested Mulberry germplasm resources based on DNJ content in mulberry leaf. (Note) germplasm resource No. are the same to those in Table 1
Conclusion
There were significant differences in the DNJ content and genetic diversity of the 36 mulberry varieties based on the SSR markers. However, cluster analyses suggested some similarities between the DNJ content and the genetic diversity of the SSR markers. This study provides invaluable information for the exploitation and breeding of mulberry varieties with a high content to fully utilize the medicinal benefits and application of DNJ.
Data Availability
All data presented in this manuscript.
References
Botton A, Barcaccia G, Cappellozza S, Tos RD, Bonghi C, Ramina A (2005) DNA fingerprinting sheds light on the origin of introduced mulberry (Morus spp.) accessions in Italy. Genet Resour Crop Evol 52(2):181–192
DongYT, Zhao EH, Gao HL, Xu YH, Zhao YJ, Fu G, Cui HJ (2018) Recent research advances of 1-deoxynojirimycin and its derivatives. China J Chin Materia Medica 43(10):1990–1997
Guo ZB, Han BM, Guo Q (2017) Genetic diversity analysis of 30 fig varieties using capillary electrophoresis detection with fluorescent SSR markers. Acta Agricul Zhejiangensis 29(9):1482–1488
Hao JY, Yi W, Yao XH, Zhao WG, Hu RZ, Cong C, Long L, Zhang DY, Wu GH, Lightfoot DA (2018) Effect of different planting areas on the chemical compositions and hypoglycemic and antioxidant activities of mulberry leaf extracts in Southern China. PLoS ONE 13(6):e0198072
Liu K, Muse SV (2005) PowerMaker: an integrated analysis environment for genetic maker analysis. Bioinformatics 21(9):2128–2129
Liu WQ, Zhu XR (2006) The determination of 1-Deoxynojirimycin (DNJ) in three parts of mulberry. Bull Seric 37(4):31–34
Liu G, Yin H, Huang GQ, Zhang JH, Wei L, Tong WH, Lin CW, Guo T (2012) Effects of balanced fertilization on mulberry leaf quality and its active ingredient content. Southwest China J Agricul Sci 25(5):1770–1776
Liu X, Chen XP, Fang XM, Fu X, Ke HT, Lv Y, Chen RF (2019) Population structure and genetic diversity of mulberry germplasm resources in Sichuan province analyzed by SSR marker. Acta Sericol Sin 45(02):165–174
Lou DS, Zou FM, Yan H, Gui ZZ (2011) Factors influencing the biosynthesis of 1-deoxynojirimycin in Morus alba L. Afr J Agric Res 6(13):2998–3006
Peng B, Hu XM, Deng W, Ye CH, Zhang H (2010) Genetic diversity analysis of mulberry germplasm resources based on SSR markers. Hubei Agric Sci 49(4):779–784
Shi XQ, Chen CJ, Li F, Li ZF, Wang ZH, Gu YY, Li HX (2013) Determination of 1-Deoxynojimycin contents in leaves of mulberry cultivars in Shandong province. Acta Sericol Sin 39(01):177–182
Tao C, Cui QY, Zhu FR, Qiu CY, Lin Q, Chen F, Chen HZ, Qu DC (2018) Difference and seasonal variation of branch DNJ content of 12 mulberry varieties planted in Guangxi. Guangxi Sericul 55(1):12–16
Vijayan K, Srivatsava PP, Nair CV, Awasthi AK, Urs SR (2006) Molecular characterization and identification of markers associated with yield traits in mulberry using ISSR markers. Plant Breed 125(3):298–301
Wang DJ, Zhao L, Wang D, Yu XF, Wei Y, Ouyang Z (2018) Transcriptome analysis and identification of key genes involved in 1-deoxynojirimycin biosynthesis of mulberry (Morus alba L.). Peer J Plant Biol 6:e5443
Yan XP, Guan S, Zhou J, Huang D (2015) An investigation to the hypoglycemic component 1-deoxynojimycin in Folium mori. Basic Clin Phar Toxicol 117:8–8
Zeng WX, Zheng S, Han L, Zhou L, Liu CY, Yu MD, Xiang ZH, Zhao AC (2018) Comprehensive evaluation of medicinal quality of mulberry leaves from 53 germplasm resources. Acta Sericol Sin 44(06):905–915
Zhang DY, Yi W, Hao JY, Hu RZ, Cong C, Yao XH, Zhao WG, Liu ZY, Long L (2018) Evaluation of the alkaloid, polyphenols, and antioxidant contents of various mulberry cultivars from different planting areas in eastern China. Ind Crops Prod 122:298–307
Zhang J, Yan XP, Li YP, Shao YY, Jiang SM, Jiang YB, Zou XY (2018) Determination and analysis on alkaloids contents in leaves of main mulberry varieties cultivated in Hunan province. Acta Sericol Sin 44(06):916–922
Zhao WG, Zhou ZH, Miao XX, Wang SB, Zhang L, Pan YL, Huang YP (2006) Genetic relatedness among cultivated and wild mulberry (Moraceae: Morus) as revealed by inter-simple sequence (ISSR) analysis in China. Can J Plant Sci 86(1):251–257
Zhao WG, Wang W, Yang YH, Huang YP, Pan YP (2008) Genetic diversity of mulberry local varieties from different ecotype as revealed by ISSR analysis in China. Acta Sericol Sin 34(1):1–5
Zhao DX, Li GC, Dong YR, Geng B, Chen CJ, Wang ZH, Lou QN, Sun JS (2019) Determination of active substances in leaves and comprehensive evaluation on medicinal quality of different mulberry hybrids. Shandong Agric Sci 51(12):100–105
Zhou S, Wang ZX, Han M (2018) Determination of 1-Deoxynojimycin contents in leaves of mulberry cultivars in Ankang city. Hubei Agric Sci 57(11):93–95123
Wang JY, Zhao YB, Feng CM (2014) Principal component analysis and cluster analysis of luffa germplasm resources in Zhejiang province. J Plant Genet Res (6):1374–1379
Yu YF, Xia YH, Huang JZ, Peng XH, Wang JW (2019) Study on the difference of mulberry bud quality in different seasons. Newsl Sericul Tea (02):1–3
Funding
This work was supported by Modern Agricultural industry Technology System of Jiangxi Province (JARS-23).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Confict of Interest
The authors declare that there is no confict of interests regarding the publication of this paper.
Ethical approval
The findings were presented without any distortion, falsification, or data manipulation, and the manuscript was not submitted elsewhere. Research poses no threat to public health or national security.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guiping, H., Xiang, C., Hongmei, C. et al. Correlation between the DNJ content and genetic diversity of SSR markers of 36 mulberry (Morus spp.) germplasm resources. Genet Resour Crop Evol 70, 1039–1047 (2023). https://doi.org/10.1007/s10722-022-01486-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01486-4