Abstract
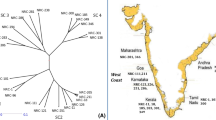
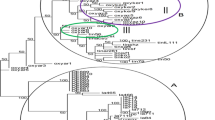
The Saccharum species members are extremely complex allopolyploids. In this study, sixty Saccharum spp. hybrids were analyzed using simple sequence repeat (SSR) primers to assess genetic diversity and population structure. The sugarcane hybrids were collected from two research stations of Pakistan and one of Sri Lanka. These were sown at the farm of Sugarcane Research Institute, Faisalabad, Pakistan. Genotyping of the hybrids with 125 polymorphic simple sequence repeat (SSR) markers amplified 476 loci. NTSYS software formed four clusters of genotypes and partitioned genotypes of three institutes into separate clusters. Maximum genetic similarity (82%) was found between CSSG-676 and CPSG-2875. Maximum genetic difference (57%) was found between CSSG-676 and SL-7103 and, thus, these genotypes showed highest level of diversity. Analysis through STRUCTURE V2.3.1 software partitioned genotypes into 3 subpopulations, which belonged to each research institute. Findings of present study indicated that SSR markers were excellent tools for genetic diversity assessment and varietal fingerprinting in Saccharum spp. hybrids. The results of this study will help to identify best sugarcane parents for planned crosses to develop elite sugarcane cultivars.




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included as Supplementary Data file.
References
Aitken KS (2021) History and development of molecular markers for sugarcane breeding. Sugar Tech. https://doi.org/10.1007/s12355-021-01000-7
Akhter S, Rauf S, Akhter B, Ghias M, Parveen N, Ibrar I, Gill SA, Ali Q (2021) Microsatellite (SSR) markers a tool; for genetic diversity assessment among sugarcane accessions. Plant Cell Biotechnol Mol Biol 22:1–7
Ali ML, Rajewski JF, Baenziger PS, Gill KS, Eskridge KM, Dweikat I (2008) Assessment of genetic diversity and relationship among a collection of US sweet sorghum germplasm by SSR markers. Mol Breed 21:497–509
Banerjee N, Siraree A, Yadav S, Kumar S, Singh J, Kumar S et al (2015) Marker–trait association study for sucrose and yield contributing traits in sugarcane (Saccharum spp. hybrid). Euphytica 205:185–201
Beckman JS, Soller M (1990) Toward a unified approach to the genetic mapping of eukaryotes based on sequence tagged microsatellite sites. Biotechnol 8:930–932
Bilal M, Saeed M, Nasir IA, Tabassum B, Zameer M, Khan A et al (2015) Association mapping of cane weight and tillers per plant in sugarcane. Biotechnol Biotechnol Equip 29:617–623
Bordonal RDO, Carvalho JLN, Lal R, de Figueiredo EB, de Oliveira BG, la Scala N (2018) Sustainability of sugarcane production in Brazil. A Rev Agron Sustain Dev 38:13
Cao K, Zhou Z, Wang Q, Guo J, Zhao P, Zhu G et al (2016) Genome-wide association study of 12 agronomic traits in peach. Nat Commun 7:13246
Cheavegatti-Gianotto A, de Abreu HMC, Arruda P, Filho JCB, Burnquist WL, Creste S et al (2011) Sugarcane (Saccharum officinarum): a reference study for the regulation of genetically modified cultivars in Brazil. Trop Plant Biol 4:62–89
Chen PH, Pan YB, Chen RK, Xu LP, Chen YQ (2009) SSR marker-based analysis of genetic relatedness among sugarcane cultivars (Saccharum spp. hybrids) from breeding programs in China and other countries. Sugar Tech 11(4):347–354
Cordeiro GM, Casu R, Mcintyre CL, Manners JM, Henry RJ (2001) Microsattellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160:1115–1123
Cordeiro GM, Pan YB, Henry RJ (2003) Sugarcane microsatellites for the assessment of genetic diversity in sugarcane germplasm. Plant Sci 165:181–189
de Morais LK, Aguiar MS, Silva PA, Câmara TMM, Cursi DE, Júnior ARF et al (2015) Breeding of sugarcane. In: Cruz VMV, Dierig DA (eds) Industrial crops: breeding for bioenergy and bioproducts. Springer, New York, pp 29–42
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Garsmeur O, Droc G, Antonise R, Grimwood J, Potier B, Aitken K et al (2018) A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Commun 9:2638
Glynn NC, McCorkle K, Comstock JC (2009) Diversity among mainland USA sugarcane cultivars examined by SSR genotyping. J Am Soc Sugarcane Technol 29:36–52
Gouy M, Rousselle Y, Chane AT, Anglade A, Royaert S, Nibouche S et al (2015) Genome wide association mapping of agromorphological and disease resistance traits in sugarcane. Euphytica 202:269–284
Guha PK, Mazumder A, Das A, Pani DR, Mondal TK (2019) In silico identification of long non-coding RNA based simple sequence repeat markers and their application in diversity analysis in rice. Gene Rep 16:100418
Hameed U, Pan YB, Muhammad K, Afghan S, Iqbal J (2012) Use of simple sequence repeat markers for DNA fingerprinting and diversity analysis of sugarcane (Saccharum spp) cultivars resistant and susceptible to red rot. Genet Mol Res 11:1195–1204
Junior CADK, Manechini JRV, Corrêa RX, Pinto ACR, da Costa JB, Favero TM, Pinto LR (2020) Genetic structure analysis in sugarcane (Saccharum spp.) using target region amplification polymorphism (TRAP) markers based on sugar-and lignin-related genes and potential application in core collection development. Sugar Tech 22(4):641–654
Lao FY, Liu R, He HY, Deng HH, Chen ZH, Chen JW et al (2008) Genetic diversity of introduced sugar cane varieties assessed by AFLP analysis. Guangdong Agric Sci 4:13–16
Li S, Liu X, Chen W, Hao Z, Bai L, Zhang D (2013) Genetic relationships among Chinese maize OPVs based on SSR markers. J Integr Agric 12:1130–1137
Li H, Rasheed A, Hickey LT, He Z (2018) Fast-forwarding genetic gain. Trends Plant Sci 23:184–186
Lima MLA, Garcia AAF, Oliveira KM, Matsuoka S, Arizono H, de Souza CLJ et al (2002) Analysis of genetic similarity detected by AFLP and coefficient of parentage among genotypes of sugar cane (Saccharum spp.). Theor Appl Genet 104:30–38
Liu K, Muse SV (2005) Power Marker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mahadevaiah C, Appunu C, Aitken K, Suresha GS, Vignesh P, Mahadeva Swamy HK, Valarmathi R, Hemaprabha G, Alagarasan G, Ram B (2021) Genomic selection in sugarcane: current status and future prospects. Front Plant Sci 12:708233
Nei N, Li W (1979) Mathematical model for studying genetic variation in terms of endonuclease. Proc Natl Acd Sci 76:5269–5273
Pan YB (2006) Highly polymorphic microsatellite DNA markers for sugarcane germplasm evaluation and variety identity testing. Sugar Tech 8(4):246–256
Pan YB (2010) Databasing molecular identities of sugarcane (Saccharum spp.) clones constructed with microsatellite (SSR) DNA markers. Am J Plant Sci 1:87–94
Pinto LR, Oliveira KM, Marconi T, Garcia AAF, Ulian EC, De Souza AP (2006) Characterization of novel sugarcane expressed sequence tag microsatellites and their comparison with genomic SSRs. Plant Breed 125:378–384
Prasad M, Varshney RK, Roy JK, Balya HS, Gupta PK (2000) The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor Appl Genet 100:584–592
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen W (2002) Documentation for STRUCTURE software: Version 2. Available from http://pritch.bsd.uchicago.edu
Rohlf FJ (1993) NTSYS-pc numerical taxonomy and multivariate analysis system, version 20. Exeter Software, Setauket, New York
Sadras VO, Slafer GA (2012) Environmental modulation of yield components in cereals: heritabilities reveal a hierarchy of phenotypic plasticities. Field Crops Res 127:215–224
Saini JK, Saini R, Tewari L (2015) Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: concepts and recent developments. 3 Biotech 5:337–353
Sharma S, Nargotra P, Sharma V, Bangotra R, Kaur M, Kapoor N et al (2021) Nanobiocatalysts for efficacious bioconversion of ionic liquid pretreated sugarcane tops biomass to biofuel. Bioresour Tech 333:125191
Silva DC, Filho LSCD, dos Santos JM, de Souza GVB, Almeida C (2012) DNA fingerprinting based on simple sequence repeat (SSR) markers in sugarcane clones from the breeding program RIDESA. Afr J Biotechnol 11:4722–4728
Singh RK, Mishra SK, Singh SP, Mishra N, Sharma ML (2010) Evaluation of microsatellite markers for genetic diversity analysis among sugarcane species and commercial hybrids. Aust J Crop Sci 4:116–125
Singh RB, Singh B, Singh RK (2019) Identification of elite Indian sugarcane varieties through DNA fingerprinting using genic microsatellite markers. Vegetos 32:547–555
Singh RB, Mahenderakar MD, Jugran AK, Singh RK, Srivastava RK (2020) Assessing genetic diversity and population structure of sugarcane cultivars, progenitor species and genera using microsatellite (SSR) markers. Gene 753:144800
Siraree A, Banerjee N, Kumar S, Khan MS, Singh PK, Kumar S, Sharma S, Singh RK, Singh J (2018) Agro-morphological description, genetic diversity and population structure of sugarcane varieties from sub-tropical India. 3 Biotech 8:469
Sleper DA, Poehlman JM (2006) Breeding field crops. Blackwell Publishing Professionals, Ames
Smiullah FAK, Aqeel A, Abdullah AI, Usman I (2013) Diversity analysis of sugarcane genotypes by microsatellite (SSR) markers. Int J Biotechnol Mol Biol Res 4:105–110
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman, San Francisco
Sukumaran S, Reynolds M, Lopes M, Crossa J (2015) Genome-wide association study for adaptation to agronomic plant density: a component of high yield potential in spring wheat. Crop Sci 55:1–11
Thirugnanasambandam PP, Hoang NV, Henry RJ (2018) The challenge of analyzing the sugarcane genome. Front Plant Sci 9:616
Vieira MLC, Almeida CB, Oliveira CA, Tacuatiá LO, Munhoz CF, Cauz-Santos LA et al (2018) Revisiting meiosis in sugarcane: chromosomal irregularities and the prevalence of bivalent configurations. Front Genet 9:213
Wang L, Qiu J, Chang L, Liu L, Li H, Pang B et al (2015) Assessment of wheat variety distinctness using SSR markers. J Integr Agric 14:1923–1935
Acknowledgements
Authors acknowledge the kind support provided by the Sugarcane Research Institute (SRI), Faisalabad, Pakistan in the form of field facilities for growing sugarcane genotypes.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
SS conducted the research and written the manuscript. MS performed all molecular analyses of the data, edited the manuscript and prepared the final draft for submission. SP, MA, IS, SS, MN, AJ, IS, UF, SM, and MZY reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shahzad, S., Saeed, M., Perveen, S. et al. Genetic diversity and population structure assessed through simple sequence repeat markers in Saccharum spp. hybrids from Pakistan and Sri Lanka. Genet Resour Crop Evol 69, 2889–2900 (2022). https://doi.org/10.1007/s10722-022-01411-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01411-9