Abstract
The Vanilla genus is a complex taxonomic group characterized by a vegetative reproduction mode combined with intra- and inter-specific hybridizations, and polyploidy events. These factors strongly impact the diversification of the genus and complicate the delimitation of taxa. Among the hundred Vanilla species, Vanilla planifolia Jacks. ex Andrews and Vanilla × tahitensis J. W. Moore are the main cultivated aromatic species. We applied Genotyping-by-Sequencing to explore the genetic diversity of these two cultivated vanilla species, seven closely related species and nineteen interspecific hybrids. The inter- and intra-specific relationships of 133 vanilla accessions were examined based on 2004 filtered SNPs. Our results showed a strong genetic structuring between the nine species studied, with wild species showing much lower heterozygosity levels than cultivated ones. Moreover, using Bayesian clustering analyses, the kinship of several hybrids could be verified. We evidenced in particular that Vanilla sotoarenasii and Vanilla odorata C.Presl may be the parental species of V. x tahitensis. The analysis of 1129 SNPs for 84 V. planifolia accessions showed a clear genetic demarcation between the vegetatively propagated traditional vanilla cultivars compared to the accessions derived from sexual reproduction, and a higher genetic diversity and lower heterozygosity of the latter (Ho = 0.206) compared to the former (Ho = 0.362). Our data are consistent with a single-step domestication for V. planifolia in accordance with the recent history of its cultivation. It also opens avenues to breed new V. planifolia varieties adapted to biotic and abiotic constraints and to reduce mutational load induced by clonal propagation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Vanilla Plumier ex Miller belongs to the Orchidaceae family and is composed of about 120 species, among which 18 (Portères 1954) to 35 (Soto Arenas 2003) are considered to bear aromatic fruits. The main cultivated species is Vanilla planifolia Jacks. ex Andrews that contributes to more than 95% of the world’s vanilla production. Native to Mesoamerican tropical forests, V. planifolia has been vegetatively propagated and cultivated in the Eastern coast of Mexico since the mid eighteenth century in response to the growing demand for vanilla pods in Europe. Vanilla cuttings were subsequently transferred into European botanical gardens, then reached the Indian Ocean region where no natural pollinator was present (Bory et al. 2008b; Lubinsky et al. 2010). The discovery in 1841 of an easy manual pollination technique by Edmond Albius in Reunion Island has led to the fast diffusion of V. planifolia into the south west Indian ocean region (Madagascar, Reunion Island, Comoros). Vanilla × tahitensis J.W.Moore, is mostly cultivated in French Polynesia and Papua New Guinea. It is supposed to have been introduced to Tahiti Island from the Philippines in 1848 (Constantin and Bois 1915). However, V. × tahitensis is no longer found in the wild and its origin has long been debated. Based on morphological characteristics, a hybrid origin between V. planifolia and Vanilla pompona Schiede (Portères 1954) or Vanilla odorata C.Presl (Portères 1954; Soto Arenas 1999) was suggested. A genetic analysis using the nuclear Internal Transcribed Spacer (ITS) and plastid DNA sequences rather suggested a hybrid origin between V. planifolia as the maternal parent and V. odorata as the paternal one (Lubinsky et al. 2008). Other aromatic species are cultivated or harvested in the wild at small scales in some localities such as V. pompona in the French Caribbean islands and V. odorata in Central and South America (Soto Arenas 1999).
Domestication, according to Martínez-Ainsworth and Tenaillon (2016), can be described as a set of consecutive stages that begins with the onset of domestication followed by an increase in the frequency of a set of desirable traits. McKey et al. (2010) highlighted the lack of knowledge on the evolutionary ecology of domesticated plants that are clonally-propagated. Vanilla is no exception and the impact of domestication on the genetics of cultivated Vanilla has received little attention. From 1793 to 1875, five introduction events of V. planifolia cuttings into Reunion Island were reported, but only one introduction in 1822 by Marchant from Europe is supposed to have been successful and to be at the origin of vanilla cultivation in Reunion Island (Bory et al. 2008b). From a historical perspective and given the very limited number of introductions, “single-step domestication” (i.e. identification of interesting genotype and direct clonal propagation) might be the rule in V. planifolia, which would generate a crop that remains close to wild progenitors (Zohary 2004). Low levels of genetic diversity are therefore expected in V. planifolia in cultivation areas such as Reunion Island, in accordance with the vegetative mode of multiplication of vanilla vines, and their recent introduction in the Indian Ocean region. Random amplified polymorphic DNA (RAPD) (Besse et al. 2004), amplified fragment length polymorphism (AFLP) (Bory et al. 2008c) and microsatellite (SSR) (Bory et al. 2008a) markers succeeded to discriminate the species and confirmed the genetic uniformity of most V. planifolia cultivars in the Indian Ocean and other cultivation areas. AFLP patterns of variation suggested that V. planifolia has evolved in introduction areas by the accumulation of point mutations through vegetative multiplication. However, these markers, and even those based on methylation patterns (MSAP) (Gigant et al. 2011), have failed to identify clusters of intraspecific genetic diversity congruent with the phenotypic variations described in cultivation (Bory et al. 2008b, c; Gigant et al. 2011). On the contrary, based on AFLP studies and linkage mapping, varieties described in V. × tahitensis were shown to result mainly from self-pollination or full sib crosses of plants belonging to the most ancient ‘Tahiti’ morphotype, with subsequent heterozygous selection (Lepers-Andrzejewski et al. 2012).
Genotyping-by-sequencing (GBS) is able to generate thousands of Single Nucleotide Polymorphisms (SNPs) markers by applying massively parallel sequencing and multiplexing methods (Elshire et al. 2011). GBS-generated SNP markers are useful to explore the genetic diversity and structure of population in order to better define phylogeny, adaptation of plants to their environment or domestication (Favre et al. 2021). Previous studies validated the efficiency of GBS to characterize Vanilla genetic diversity and to identify hybrids (Hu et al. 2019; Alomia et al. 2021). Herein, we developed SNP markers derived from GBS data to study intraspecific diversity and enlighten the evolutionary history of cultivated vanilla in its introduction areas. Our study focused on genetic diversity of Vanilla resources conserved in the Biological Resources Centers (BRCs) Vatel and Etablissement Vanille de Tahiti (EVT) (Roux-Cuvelier et al. 2021): cultivated V. planifolia and V. × tahitensis species, seven wild relatives originating from tropical America (Soto Arenas 2003) and selfed-progenies of V. planifolia and interspecific hybrids. This well characterized germplasm constitutes a material of choice to assess the genome-wide genetic diversity, the impact of domestication processes and breeding on genetic diversity levels in cultivated Vanilla compared to wild genotypes.
Materials and methods
Plant material and DNA extraction
A panel of 137 Vanilla spp. accessions was used for GBS sequencing, including (i) the two cultivated species V. planifolia (88 accessions) and V. × tahitensis (7 accessions), (ii) 7 closely related species V. odorata (8 accessions), Vanilla cribbiana Soto Arenas (4 accessions), V. sotoarenasii (4 accessions), Vanilla insignis Ames (2 accessions), V. pompona (2 accessions), Vanilla bahiana Hoehne (2 accessions), Vanilla helleri A.D. Hawkes (1 accession) and (iii) 19 interspecific hybrids (Supporting Information Table S1). All accessions were grown in shade house or in vitro in Reunion Island and French Polynesia and were selected from a large collection of over 700 accessions conserved in the French BRCs Vatel (CIRAD, Reunion Island) and EVT (Raiatea) (Roux-Cuvelier et al. 2021). Accessions were selected in order to maximize variability for origin, variety and ploidy level and better evaluate the species diversity. Traditional cultivars of V. planifolia and V. × tahitensis collected in fields were classified as vegetatively (asexually) propagated clones or “cuttings”. Accessions obtained by sexual reproduction (selfed-progenies or intra-specific hybrids) were classified as “seedlings”. All accessions were clonally propagated by cuttings or micro cuttings in vitro for their conservation in BRCs Vatel and EVT (Supporting informations S1 and S2). High molecular weight DNA of each accession was extracted from 25 mg of lyophilized young leaves using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Genomic DNA was quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and normalized at 50 ng/µL. DNA homogeneity and quality was assessed by enzymatic digestion with HindIII (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and run in a 2% agarose gel.
Library preparation and sequencing
Library preparation was performed by the Regional genotyping technology platform (UMR AGAP, CIRAD, Montpellier, France) as described by Elshire et al. (2011). Two kinds of adapters are used for constructing GBS libraries, a common adapter and a sample-specific barcode adapter. Both adapters are designed to fit with Illumina sequencing. Adapters were mixed together in a 1:1 ratio and plated into two 96-well plates, so that each well contained one specific barcode. Extracted DNA was added into the 96-well plates and digested with PstI methylation-sensitive restriction enzyme (New England Biolabs, Ipswich, Massachusetts, USA). Adapters were ligated to the ends of the DNA fragments using a T4 ligase (New England Biolabs, Ipswich, Massachusetts, USA). Samples were then pooled together, amplified by Polymerase Chain Reaction (PCR) and purified to remove unreacted adapters. The GBS library was sequenced on Illumina HiSeq3000 sequencer (Illumina Inc., San Diego, California, USA) with DNA-seq single-read protocol at the GeT-PlaGe platform (INRAE, Toulouse, France).
Sequence analysis and SNP calling
Sequence quality was checked with FastQC (Andrews 2010). Low-quality reads, reads with uncalled bases and reads with Illumina adapter sequences were removed using the Cutadapt software (Martin 2011). The remaining reads were assigned to each sample using the GBS barcode splitter tool (https://sourceforge.net/projects/gbsbarcode/). Demultiplexed sequences were trimmed to 140 bp to normalize the length between individuals. SNP calling was performed using STACKS de novo pipeline (Catchen et al. 2013) and identified SNPs were converted into Variant Call Format file (VCF; Danecek et al. 2011). Low-quality SNPs were filtered out with vcfR package 1.11.0 (Knaus and Grünwald 2017) from Rstudio (version 3.6.3) (R Development Core Team 2010) and using successive filters: minimum minor allele frequency < 10%, missing data per site > 30% and up to 3 SNPs per locus. SNPs with an allele frequency below 10% were discarded because these very rare variants probably resulted from genotyping errors, while retaining rare alleles that are associated to under-represented samples in the dataset. The V. planifolia accessions were used to study structure within such a clonally propagated vanilla, and only polymorphic SNP markers were kept. Loci with unfiltered SNP markers and filtered SNPs were mapped over the Daphna V. planifolia chromosomes (Hasing et al. 2020) to check SNP distribution and density. The genotyping data sets were converted into biallelic tables by vcfR package (Knaus and Grünwald 2017), which were used for both phylogenetic relationships and population structure analysis.
Genetic diversity and phylogenetic analysis based on SNP markers
The number of effective alleles (Ne), Shannon’s information index (I), observed heterozygosity (Ho) and percentage of polymorphic loci (P) were calculated using GenAIEx (version 6.502) (Peakall and Smouse 2012). The values were compared across the nine species and interspecific hybrids using the complete genotyping data set, and across different types of V. planifolia cultivars. From the complete genotyping dataset, a dissimilarity coefficient was calculated with DarWIN software (Perrier and Jacquemoud-Collet 2006) using the simple matching index (Sokal and Michener 1958): \({d}_{ij}=1-\frac{1}{L} {\sum }_{l = 1}^{L}\frac{{m}_{l}}{\pi }\) where \({d}_{ij}\) is the dissimilarity between units i and j; L the number of loci; \({m}_{l}\) the number of matching alleles for locus l; and π the ploidy. Distance trees were constructed from 1000 bootstrap replicates using the Unweighted Neighbor-Joining method (Saitou and Nei 1987). Trees were then converted into Phylip file and plotted with FigTree software (Rambaut 2006).
Population structure analyses
Principal coordinates analyses (PCoA) were performed using the complete genotyping data set and the V. planifolia data set with GenAIEx (version 6.502) (Peakall and Smouse 2012). The population structure was analyzed to identify clusters of genetically related individuals using the Bayesian clustering method implemented in STRUCTURE (version 2.3.4) (Pritchard et al. 2000). The STRUCTURE analysis was first performed between V. odorata, V. planifolia, V. pompona, V. × tahitensis and hybrids, and then between all the V. planifolia accessions. The admixture model of STRUCTURE was chosen on the assumption that each individual had ancestry from one or more of K genetically distinct sources. Ten independent runs were performed for each K from K = 1 to K = 10, with a burn-in period of 10,000 and 100,000 Markov-chain Monte Carlo (MCMC) iterations after burn-in. The best number of K was chosen with the ΔK method (Evanno et al. 2005) by running the STRUCTURE HARVESTER (Earl and vonHoldt 2012). For STRUCTURE analysis within V. planifolia, accessions were assigned to one cluster if their probability of belonging to this cluster is higher than 60%.
Results
Sequencing and SNP calling
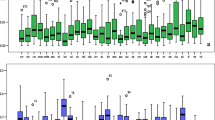
The sequencing of GBS libraries resulted in 1,143,846,935 single-reads of 150 bp for 137 Vanilla accessions (Supporting information Tables S1, S2, S3 and S4). After the cleaning step, 554,565,581 reads of 140 bp were demultiplexed for each individual (51.5% of the reads were discarded). CR0026, CR0151 and CR2564 samples, with a low sequencing depth (0; 6377 and 2657 reads, respectively), were removed from the study. The de novo pipeline reconstructed 828,215 loci including 81,987 identified as biallelic (10.3%) with 225,857 raw SNPs. Among the 80,203 loci with a sequencing depth ≥ 5 reads, 32% mapped on the 14 chromosomes of the published V. planifolia Daphna cv genome (Hasing et al. 2020) (Supporting information Fig. S1). The 194,625 SNPs distributed on these loci were filtered out based on: minimum minor allele frequency (MAF) < 10%, missing data per site > 30%, and a maximum of 3 SNPs per locus. The data set obtained consisted of 2040 high-quality filtered SNPs. The accession V. planifolia CR0844 was removed due to a missing data rate > 45%. The final data set consisted of 133 genotyped vanilla samples using 2004 filtered SNPs, with a mean heterozygosity of 19.4% (± 16.7%) and a mean missing data rate of 19.5% (± 6.3%). A specific subset of 84 V. planifolia individuals using 1129 SNPs was produced from the complete genotyping matrix using the same filters, with a mean heterozygosity of 43.6% (± 14.9%) and a mean missing data rate of 15.2% (± 7.5%). Among the 2004 and 1129 SNPs, 1916 (95.6%) and 1081 (95.8%) mapped onto the 14 chromosomes of the V. planifolia cv Daphna genome. For the 2004 SNP matrix, the density of SNPs per chromosome ranged from one SNP every 527 kb (chromosome 2) to one SNP every 230 kb (chromosome 1), with an average of one SNP every 359 kb (Fig. 1a). For the 1129 SNP matrix, the lowest density of SNPs was detected on chromosome 3 (one SNP every 1024 kb) and the highest on chromosome 1 (one SNP every 429 kb), with an average of one SNP every 641 kb (Fig. 1b).
Genotyping-By-Sequencing SNP distribution and density on V. planifolia cv Daphna chromosomes. a From 2004 filtered SNPs dataset. b From 1129 filtered SNPs dataset. Horizontal axis displays the chromosome length. The density scale indicates the number of SNPs within 500 Kb window size. The plot shows the distribution of GBS SNPs across the 14 chromosomes
Genetic relationships across species within the genus Vanilla
The unweighted Neighbor-Joining (NJ) tree built with the complete genotyping matrix revealed two major groups statistically supported with bootstrap values equal to 1. On one side of the tree, a group comprised the V. planifolia accessions plus six hybrids, and on the other side, a group comprised the accessions from the wild relative species (V. bahiana, V. cribbiana, V. helleri, V. insignis, V. odorata and V. pompona) (Fig. 2). V. × tahitensis accessions and most of the interspecific hybrids branched in the tree in intermediate positions between the two major groups, with bootstrap values ranging from 0.6 to 1. V. sotoarenasii accessions were closer to the V. planifolia group than to wild relatives but were clearly individualized (bootstrap value = 1). The wild relative species group was divided into four subgroups statistically supported by bootstrap values > 0.9. The first subgroup comprised V. pompona, the second subgroup comprised V. cribbiana, the third subgroup comprised V. odorata accessions, V. helleri CR3614 and V. insignis cf. CR2688, and the last one comprised V. bahiana accessions, V. insignis cf. CR0087 and two hybrids having at least one V. bahiana parent. PCoA analyses showed a similar clustering of the accessions (Supporting information Fig. S2). The first coordinate of the PCoA explained 44.49% of the genetic variability and separated V. planifolia from the wild species, with hybrids in intermediate position. The second coordinate, explaining 7.03% of the genetic variability, separated V. × tahitensis from the other accessions. Genome-wide heterozygosity (Ho) per species was calculated using the complete genotyping matrix (Table 1). Ho values were of similar range in the cultivated species, V. planifolia (0.287 ± 0.001) and V. × tahitensis (0.239 ± 0.009), and on average a hundred times higher than the Ho values observed in wild relative species (ranging from 0.001 ± 0.000 in V. cribbiana to 0.031 ± 0.002 in V. sotoarenasii). The mean Ho of vegetatively propagated V. planifolia (0.362 ± 0.008) was significantly higher than the mean Ho in selfed-progenies (0.206 ± 0.005). Hybrids revealed Ho levels (0.246 ± 0.004) close to those observed in cultivated vanilla. Shannon’s information index (I) was highest for interspecific hybrids (0.551 ± 0.551), followed by V. planifolia and V. × tahitensis (0.374 ± 0.008 and 0.205 ± 0.007 respectively). Among the wild species, I values ranged from 0.001 ± 0.001 in V. helleri to 0.110 ± 0.005 in V. odorata. The percentage of polymorphic SNPs was higher in the cultivated vanillas and in interspecific hybrids (> 30%), compared to wild relatives (< 23%).
Phylogenetic structuration between cultivated vanillas and wild relative species. Unweighted Neighbor-Joining tree constructed from 1000 bootstrap replicates using 2004 SNPs and 133 accessions. Bootstraps values higher than 0.5 are shown between V. planifolia, V. × tahitensis, wild species and hybrids. Scale bar shows genetic distance. *V. planifolia accessions with unknown reproduction mode
Genetic relationships between V. planifolia and several of its hybrids
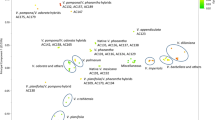
In the light of our GBS-generated SNPs we explored the relationships between several species and derived interspecific hybrids. A Bayesian clustering analysis based on 2004 SNPs (Fig. 3a) was performed on a reduced dataset including six hybrids, two V. pompona (Fig. 3b), seven V. odorata (Fig. 3c), seven V. × tahitensis (Fig. 3d), four V. sotoarenasii (Fig. 3e), and a representative subset of 12 V. planifolia accessions (Fig. 3f) selected among subgroups identified in Fig. 2. The estimated likelihood was greatest for K = 4, suggesting the presence of 4 clusters corresponding to the 4 species V. odorata, V. planifolia, V. pompona and V. sotoarenasii. For supposed first-generation hybrids, we were expecting a probability of 0.50 for each parental genome assignment. The seven V. × tahitensis accessions had in average 56.5% of their SNPs attributed to V. sotoarenasii and 43.5% to V. odorata (X-squared = 1.69, df = 1, p-value = 0.1936), confirming the interspecific hybrid status of this species. The accession CR1415 (CR0017 x CR0017) had similar proportions in its genome (53.6/46.4%) than its parent V. × tahitensis CR0017 (55.9/44.1%). For the V. pompona × V. sotoarenasii hybrid CR1156, 61.4% of the SNPs were assigned to V. sotoarenasii and 38.6% to V. pompona. These proportions did not fit the hypothesis of first-generation hybrid (X-squared = 5.20, df = 1, p-value = 0.0226). The SNPs of hybrid CR1725 were assigned to V. pompona (67.6%) and V. planifolia (32.4%) which was also inconsistent with hypothesis of 0.50/0.50 distribution of parental genomes (X-squared = 12.39, df = 1, p-value = 0.0004). The SNPs of hybrid CR0139 were assigned to V. planifolia (54.2%), V. pompona (38.0%) and V. sotoarenasii (7.80%). Therefore, these three first-generation hybrids showed parental inheritances of SNPs significantly deviating from the hypothetical ratio of 0.5/0.5.
Origin of V. × tahitensis shown by bayesian clustering and comparison of morphological traits. a Population structure of 38 vanilla accessions using 2004 significant SNPs. Colours represent different assigned clusters. The X-axis provides accession and species names and the y-axis provides the probability of each accession belonging to the assigned cluster. Front view of entire flowers of b V. pompona CR0018, c V. odorata CR0116, d V. × tahitensis CR0017, e V. sotoarenasii CR0068 and f V. planifolia CR0040
For back-cross hybrids, we were expecting 0.75/0.25 distribution of the parental genomes. The hybrid CR2717 ((V. planifolia × V. pompona) × V. planifolia) had 79.1% of its genome assigned to V. planifolia and 20.8% to V. pompona, which was consistent with back-cross ratio hypothesis (X-squared = 0.93, df = 1, p-value = 0.3347). The hybrid CR2718, which is a cutting of CR2717 showed similar results (79.7% assigned to V. planifolia and 20.2% to V. pompona, and X-squared = 1.22, df = 1, p-value = 0.2699). The hybrid CR0003, that comes from a V. planifolia × V. × tahitensis cross, revealed ancestry from V. planifolia (60.9%), V. odorata (27.6%) and V. sotoarenasii (11.5%) as expected, but the proportions (60.9/27.6/11.5%) were inconsistent with the 50/25/25% proportions expected for such a cross (X-squared = 9.94, df = 2, p-value = 0.0070).
Intraspecific genetic structuration within V. planifolia
The V. planifolia accessions derived from sexual reproduction showed some structuration in the NJ tree (Fig. 2) with subgroups supported by bootstrap values equal to 1, whereas no genetic structuration was supported by bootstraps values in vegetatively propagated accessions. Among the accessions with unknown reproduction mode, CR2093, ‘Colibri’ CR2687, CR2100 and CR2102 branched within cuttings, while CR0510, CR0196 and CR0628 (‘Aiguille’) branched within seedlings. The tetraploids ‘Grosse Vanille’ CR0802 and CR0641 and the triploids ‘Sterile’ CR0645 and CR0630, were grouped together with cuttings without significant structuration between these accessions. The cultivar ‘Petite Mexique’ CR0632, supposed to derive from clonal propagation, was branched with accessions derived from sexual reproduction.
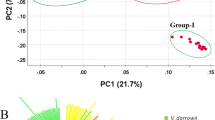
The first and second coordinates of the PCoA of V. planifolia accessions explained 13.92% and 8.29% of the genetic variability, respectively (Supporting information Fig. S3a). These values were lower than those observed from the complete dataset PCoA (Supporting information Fig. S2), indicating a low structuration within the V. planifolia group. Individuals obtained by vegetative propagation were clustered along the first axis, while individuals derived from sexual reproduction were much more dispersed throughout the plan. No structuration related to geographic origin or the ploidy level of cultivars could be evidenced by PCoA (Supporting information Fig. S3b and c).
Genetic structure was explored by a Bayesian clustering analysis with 84 V. planifolia accessions using the 1129 SNP matrix (Supporting information Fig. S4a). The estimated likelihood was the greatest for K = 2, K = 3 and K = 4, suggesting the presence of 2 to 4 genetic clusters (Supporting information Fig. S4b and c). Accessions were assigned to one specific cluster if their probability of belonging to this cluster was higher than 60%, in order to facilitate interpretation based on a majority rule basis. If not, they were considered admixed. For K = 2, the majority (84.21%) of the accessions derived from vegetative propagation were assigned to cluster K2_2, whereas 84.62% of the accessions derived by sexual reproduction were assigned to cluster K2_1 (Fig. 4a). For K = 4, 76.31% of the accessions derived from vegetative propagation were assigned to the cluster K4_2, whereas admixture and a greater diversity of clusters were observed in accessions derived from sexual reproduction (Fig. 4b). Among unknown accessions, five were assigned to the ‘vegetative’ clusters (K2_2 and K4_2), one to the ‘sexual’ clusters (K2_1 and K4_1), and one appeared admixed for K = 4 (Supporting information Fig. S4a).
Genetic structure of 84 V. planifolia using 1129 informative SNPs. a Proportion of accessions assigned to one of the two clusters determined by STRUCTURE for K = 2, according to mode of reproduction. b Proportion of accessions assigned to one of the four clusters determined by STRUCTURE for K = 4, according to mode of reproduction. The dotted ground indicates accessions classified as admixed
Discussion
Our study confirmed that GBS is a powerful genomic tool for the identification of highly informative SNPs for the study of the inter- and intra-specific genetic diversity in the Vanilla genus. Here, we applied GBS to explore the genetic relationships and genetic structure of the two cultivated Vanilla species, seven closely related species, and 19 interspecific hybrids from the well documented vanilla collections maintained ex situ at BRCs Vatel and Vanille de Tahiti. GBS genotyping yielded 2004 high quality filtered SNPs for the Vanilla genus, among those a subset of 1129 SNPs was used for V. planifolia. The majority of the SNPs (95%) successfully mapped on the published V. planifolia cv Daphna genome (Hasing et al. 2020), except for the extremity of the chromosome 2 not covered with GBS-generated SNPs. This might result from erroneous chromosome 2 assembly in the Daphna genome or a richness in repetitive elements and low-complexity regions in this chromosome part.
Our GBS data supported the current taxonomy for most of the species studied (Bouetard et al. 2010). As expected, V. planifolia and wild relative species were strongly separated. Hybrids from crosses between V. planifolia and a wild species were in intermediate position on the NJ tree or close to the V. planifolia parent. The V. pompona group was the most distant from the V. planifolia group in agreement with plastid DNA analysis (Bouetard et al. 2010). Accessions attributed to the species V. odorata formed a large and structured group, except CR3612 which probably has a hybrid origin. Indeed, morphology of flowers and fruits obtained recently for CR3612 suggested a kinship with a species close to V. pompona, while the long and narrow leaves were related to V. odorata (Supporting information Fig. S5a). V. odorata cf. CR2686 was probably misidentified and should rather be classified within V. insignis, since it is genetically close to V. insignis CR2688 (Fig. 2) and showed broad leaves and rough stem typical of this species. This would make V. insignis a very close but nevertheless distinct group (bootstrap = 1) from V. odorata. Accession CR0087 identified as V. insignis cf. showed proximity in the NJ tree to the V. bahiana group and is very close to its hybrid with V. bahiana. The flowers of CR0087 differ from those of V. bahiana by their slightly larger size but above all by the presence of very developed orange papillae on the lip of the labellum (Supporting information Fig. S5b). The V. helleri accession CR3614, nested within the V. odorata group, could be conspecific to this species. The V. × tahitensis accessions formed a large and structured group close to, but clearly distinct from its hybrids and from the V. planifolia group, contrary to what was observed with plastid DNA analysis (Bouetard et al. 2010).
The V. sotoarenasii group was closely related but distinct from V. planifolia (bootstrap value = 1), as previously established (Bory et al. 2008c; Bouetard et al. 2010; Azofeifa-Bolaños et al. 2017). V. sotoarenasii did not appear like a hybrid in the STRUCTURE analyses, but like a distinct genetic group with specific SNPs. In a commendable effort to limit specific inflation in the genus Vanilla, it was recently proposed that V. sotoarenasii is conspecific to V. planifolia (Karremans et al. 2020). The authors based their proposal on the absence of genetic differences between the two species and argue that the morphological variations of vanilla vines in the Cahuita area (Costa Rica), where large populations of V. sotoarenasii have been described (Azofeifa-Bolaños et al. 2017), do not allow to distinguish one species from the other. These two arguments do not hold. First, as indicated previously (Azofeifa-Bolaños et al. 2017), a small (2 nucleotides) but steady difference separates the ITS sequences of V. sotoarenasii from those of V. planifolia. The GBS data presented here, based on 2004 SNPs, provided clear evidence of genetic differentiation between V. planifolia and V. sotoarenasii. Second, the argument that the morphological variability of V. planifolia contains that of V. sotoarenasii is not supported by any quantitative data. On the contrary, the photograph of numerous fruits produced by the authors (Karremans et al. 2020) to show the wide variability in V. sotoarenasii demonstrated that the fruits of V. sotoarenasii were always between 9 and 15 cm in length, and indehiscent, which is much less than the 21 cm average length of V. planifolia fruits, that are moreover very predominantly dehiscent (Díaz-Bautista et al. 2018). In addition, the variations in shape and color of the fruits are much more certainly due to different stages of maturity, and incomplete natural pollinations, than to phenotypic plasticity that remains to be demonstrated. Similarly, variations in leaf shape and color are common in vanilla plants in relation to biotic and abiotic factors during their growth and have little taxonomic value (Soto Arenas and Cribb 2013). The other objection of these authors that the flower size measurements of V. sotoarenasii (Azofeifa-Bolaños et al. 2017) would be biased by the fact that they were made on an accession grown under controlled conditions is not supported by any measurement of variability in wild populations. Unlike vegetative organs, the morphology of reproductive organs is little impacted by environmental conditions and therefore has a better taxonomic value. On the other hand, as mentioned previously (Azofeifa-Bolaños et al. 2017) and documented recently in V. pompona (Watteyn et al. 2022) flower size is, among others, an important trait affecting reproductive isolation. In the case of difficult groups such as the genus Vanilla, alpha-taxonomy is often not very discriminating or even risky, and an integrative taxonomy approach (Andriamihaja et al. 2022) allows more effectively to dissect the relationships between closely related taxa. In this perspective, the bundle of genetic, morphologic and ecologic arguments clearly plead in favor of the recognition of V. sotoarenasii as a valid species which might have recently evolved from V. planifolia by geographic and/or reproductive isolation. However, the hypothesis of a hybrid origin of V. sotoarenasii involving V. planifolia and a species not included in our study, cannot be completely ruled out.
Bayesian clustering analyses based on GBS-generated SNPs proved to be a powerful tool to assess parental genetic contributions of hybrids, in case of lack of information on a genetic resource, mislabelling or uncontrolled pollination. In this study, we confirmed, or infirmed the taxonomic position and kinship for several species and hybrids. However, some first-generation hybrids did not show the expected 50/50 parental assignment probabilities. Most of the hybrids assessed have V. pompona as parental species, which is known to have a much larger genome than V. planifolia (2C = 8.18 to 10.72 pg) (Bory 2007). The observed deviations could therefore be explained by parental genome complexity. For instance, molecular cytogenetics studies on polyploid sugarcane cultivars, deriving from a few interspecific hybridization events performed a century ago by breeders, highlighted an uneven contribution of each parental genome, with 75–85% of their chromosome originating from one parental species (S. officinarum) and 15–25% from the other parent (S. spontaneum), with some chromosomes derived from interspecific recombination (Piperidis and D’Hont 2020). A recent GBS study (Alomia et al. 2021) also included some V. planifolia x V. pompona hybrids, but the parental contributions were unfortunately not quantified, preventing any comparison. Nevertheless, GBS based Bayesian clustering enabled us to undoubtedly identify the parental origin of all the hybrids studied. Our results open important questions about possible genome rearrangement in interspecific crosses between relatively distant species in the genus Vanilla, that will need to be addressed further.
Previous assessment of V. × tahitensis origin, based on nuclear (ITS, GBS) and plastid (rbcL) loci, suggested a hybrid origin between V. planifolia and V. odorata (Lubinsky et al. 2008; Hasing et al. 2020; Alomia et al. 2021). Our GBS data confirmed at the genomic level (whole genome scale) the hybrid origin of Tahitian Vanilla evidenced in Lubinsky et al. (2008) study, and the proportions of genetic parental contributions (close to 0.50 for all accessions) were compatible with the hypothesis of a first-generation hybrid. However, V. sotoarenasii material was missing in Lubinsky et al.’s work. We contributed to clarify the question of the origin of V. × tahitensis by including for the first time well characterized V. sotoarenasii accessions and genomic data. According to our analysis, V. odorata is indeed one parent, but the second parent is closer to V. sotoarenasii than to V. planifolia as previously stated. The flower morphology of V. × tahitensis (Fig. 3c), with traits that are close to V. odorata (Fig. 3b) and to V. sotoarenasii (Karremans et al. 2020), supports this observation (Fig. 3d). Alomia et al. (2021) identified, by GBS, V. planifolia wild accessions from Belize (referred to as Type 2) as the possible true parent of V. × tahitensis. These accessions appeared very close genetically to V. sotoarenasii (Alomia et al. 2021). This observation reinforces our hypothesis and we suggest that the Type 2 V. planifolia from Belize in Alomia et al. (2021) could possibly be V. sotoarenasii. Recent data from Chambers et al. (2021) indeed showed that Type 2 V. planifolia and V. sotoarenasii cannot be separated genetically in PCA and Structure analyses. A morphological characterization of these Belize accessions could also allow to further verify this hypothesis. Karremans et al. (2020) have also argued that V. sotoarenasii is not a species at all, but most likely an introgressed hybrid involving V. odorata, V. × tahitensis, and/or V. planifolia. Our results clearly rather show that V. sotoarenasii is a species close to V. planifolia, and that V. × tahitensis is a hybrid between V. sotoarenasii and V. odorata. Nevertheless, even if V. sotoarenasii was dismissed as a species as argued by Karremans et al. (2020), although we do not agree with this, the maternal origin of V. × tahitensis was more precisely addressed by our study and should be searched for in V. sotoarenasii-like populations. According to low divergence of ITS sequence data, V. × tahitensis appeared to be evolutionarily recent and it was suggested that it resulted from natural or man-mediated pollination in Mesoamerica (Lubinsky et al. 2008). V. sotoarenasii is present in southwestern Costa Rica (Azofeifa-Bolaños et al. 2017), and may also be also present in the north of Costa Rica. It was observed in northeastern Colombia (Choco, MG personal observation) and if it is confirmed that it is also present in Belize, this would suggest that the geographic range of V. sotoarenasii is much wider in America than the Caribbean area of Costa Rica where it has been reported so far. The other parent of V. × tahitensis, V. odorata, has a large distribution area covering Central and tropical South America. Thus, the hybrid origin of V. × tahitensis is compatible with the sympatric range of both parents. Historically, the species is said to have been introduced from America to French Polynesia via the Philippines by Amiral Hamelin in 1848 (Correll 1953; Portères 195). The history of migrations and exchanges between the Viceroyalty of New Spain, the Philippines and Pacific islands between the sixteenth and eighteenth centuries (Merrill 1954) is compatible with this hypothesis. Indeed, it can be suggested that the shipment of pods or cuttings could have occurred 300 years ago on board of the Manila Galleons, the first Spanish ships that crossed the Pacific Ocean and introduced many plants from America to Asia and Oceania (Merrill 1954; Lubinsky et al. 2008).
Genome-wide Ho levels in wild species were very low (Ho = 0.001 to 0.031) compared to cultivated species (Ho = 0.287 and Ho = 0.239), suggesting frequent inbreeding in the wild. Although vanilla flowers possess a rostellum preventing self-pollination, most species studied are self-compatible and selfing can occur via geitonogamy between different flowers from the same individual (Gigant et al. 2016). Further population genetic studies are needed to precisely assess possible genetic threats such as inbreeding on these species in the wild. The very low Ho in wild species can also be shown when observing GBS data in Alomia et al. (2021) and Hu et al. (2019), although the authors did not discuss this result. V. planifolia in cultivation was shown to have very low levels of diversity (AFLP, RAPD, SSR) (Besse et al. 2004; Bory et al. 2008a, c), and a single introduction in the south western Indian ocean area was suggested (Bory et al. 2008c). This pattern, compatible with a single-step domestication, was confirmed for V. planifolia using GBS. As clonal propagation is known to increase heterozygosity by the accumulation of point mutations (Balloux et al. 2003), single-step domestication process is always associated with high levels of heterozygosity, as shown for cassava (Elias et al. 2004) and hops (Jakše et al. 2001). Long established clones were indeed highly heterozygous as shown in cassava landraces (Ho = 0.50 to 0.71) (Pujol et al. 2005). The high genome-wide Ho levels detected (0.362) in cultivated clonal V. planifolia varieties worldwide are in accordance with this hypothesis. Most vanilla cultivars indeed result from almost 200 years of intense clonal propagation of the cuttings initially introduced to La Reunion in 1822. The present values are indeed high compared to those reported in V. planifolia wild populations using 15 allozyme loci (Ho = 0 to 0.078) (Soto Arenas 1999). They are higher than those revealed by 14 SSR markers (Ho = 0,154) (Bory et al. 2008a), but close to those estimated from ALFP (Ho = 0,295) (Bory et al. 2008c) in a similar set of V. planifolia varieties. SSR markers’ length evolves more quickly than point mutation, and SSR were shown to reveal higher Ho than SNPs in population genetics studies (Fischer et al. 2017). However, in our particular case, if the levels of heterozygosity are due to the accumulation of mutations during clonal propagation (rather than from demographic and reproductive history as in natural populations), only SNPs can detect such mutations, since SSR only assess length variations in the number of microsatellite repeats. The Ho value obtained from dominant AFLP markers was deduced from the proportions of segregating bands in self-progenies (Bory et al. 2008c) and appears slightly underestimated. Our Ho levels are much higher than those revealed by Hu et al (2019) (Ho = 0.0322 to 0.0457) but this might be due to their SNP filtration of Ho > 0,2 (Hu et al. 2019) because the same accessions studied in a recent GBS analysis (Alomia et al. 2021) showed much higher Ho levels, compatible with our results. Interestingly, we therefore demonstrate here, thanks to a well characterized set of accessions from BRC Vatel, that Ho levels can be used to differentiate wild from cultivated vanillas. This could be very useful for less characterized collections. Indeed, this might indicate that the “hidden diversity” detected in V. planifolia by Alomia et al. (2021) using GBS could rather simply correspond to cultivated (Ho > 0.20, Type 1) compared to wild (Ho < 0,03, Type 2) accessions. Type 3 accessions with intermediate Ho values (0.08 to 0.15) could be cultivars naturalized in the wild, or wild accessions with high natural cloning rate.
Based on AFLP analysis, V. × tahitensis was suggested to have a different domestication history than V. planifolia (Gigant et al. 2011; Lepers-Andrzejewski et al. 2011, 2012). Although a single introduction origin was also suggested, it has been followed by one or two generations of self-pollination, as shown by the detection of recombination events using AFLP graphical genotypes (Lepers-Andrzejewski et al. 2012). V. × tahitensis therefore rather fits the pattern of a single-step domestication (one introduction) followed by subsequent recombination-and-selection cycles (McKey et al. 2010). This hypothesis is supported by genome-wide Ho that is lower in V. × tahitensis (Ho = 0.239) than in V. planifolia (Ho = 0.362).
Analysis of 1129 SNPs for 84 V. planifolia accessions showed a clear demarcation between the vegetatively propagated traditional vanilla cultivars compared to the accessions derived from sexual reproduction. GBS-generated SNPs could be used efficiently to better define the origin of unknown accessions: CR0196, CR2093, CR2100, CR2102 and ‘Colibri’ CR2687 were assigned to vegetatively propagated accessions, CR510 was assigned to accessions derived from sexual reproduction and CR0628 remains unknown. The cultivar ‘Petite Mexique’ CR0632 supposed to be a traditional cultivar with clonal propagation was branched and clustered with accessions derived from sexual reproduction.
Selfed-progenies and intra-specific crosses in V. planifolia showed an increased level of diversity (Fig. 4, Supporting information Fig. S3a, S4) as previously suggested by AFLP study (Bory et al. 2008c). They also showed a reduced genome-wide heterozygosity (Ho = 0.206) as compared to the original parental cuttings group (Ho = 0.362), as expected following selfing. Although heterozygosity is a favorable trait (ie hybrid vigor), the heterozygosity born from strict clonal propagation leads to the accumulation of deleterious mutations (McKey et al. 2010). Therefore V. planifolia plantations sustainability could be threatened by this mutational load (McKey et al. 2010). To preserve the adaptive potential of V. planifolia, the maintenance of mixed clonal/sexual systems is considered the best strategy (McKey et al. 2010). Efforts that have been engaged at BRCs Vatel and EVT to create new varieties from selfing will contribute to increase diversity levels, and also to reduce heterozygosity levels and therefore release mutational load. This has been a very successful strategy to create Fusarium resistant plant (Handa) (Grisoni and Dijoux 2017) from the selfing of a V. planifolia parent. Our study clarified the origin of Vanilla spp. from a large collection and their genetic diversity and structure, and provides new informations for breeding programs that contribute to the enhancement and protection of these materials. Tahitian vanilla, known to be more fragrant and fruity, illustrates the potential of hybridization to select new varieties.
Data availability
The raw sequence data have been deposited in the Short Read Archive under NCBI BioProject ID PRJNA756864. Biological samples used in this study have been deposited in the BioSamples Database under the accession identifier SAMN 20,927,883 to 20,928,015, and SAMN20981991 to SAMN20981994.
References
Alomia YA, Chambers A, Brym M et al (2021) Genotyping-by-sequencing diversity analysis of international Vanilla collections uncovers hidden diversity and enables plant improvement. Plant Sci. https://doi.org/10.1016/j.plantsci.2021.111019
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Andriamihaja CF, Botomanga A, Misandeau C, et al (2022) Integrative taxonomy and phylogeny of leafless Vanilla orchids from the South West Indian ocean region reveal two new Malagasy species. J Syst Evol
Azofeifa-Bolaños JB, Gigant LR, Nicolás-García M, et al (2017) A new vanilla species from Costa Rica closely related to V. planifolia (Orchidaceae). EJT. https://doi.org/10.5852/ejt.2017.284
Balloux F, Lehmann L, de Meeûs T (2003) The population genetics of clonal and partially clonal diploids. Genetics 164:1635
Besse P, Silva DD, Bory S et al (2004) RAPD genetic diversity in cultivated vanilla: Vanilla planifolia, and relationships with V. tahitensis and V. pompona. Plant Sci 167:379–385. https://doi.org/10.1016/j.plantsci.2004.04.007
Bory S (2007) Diversity of Vanilla planifolia in the Indian ocean and its related species : genetics, cytogenetics and epigenetics aspect. Université de La Réunion
Bory S, Da Silva D, Risterucci A-M et al (2008a) Development of microsatellite markers in cultivated vanilla: polymorphism and transferability to other vanilla species. Sci Hortic 115:420–425. https://doi.org/10.1016/j.scienta.2007.10.020
Bory S, Grisoni M, Duval M-F, Besse P (2008b) Biodiversity and preservation of vanilla: present state of knowledge. Genet Resour Crop Evol 55:551–571. https://doi.org/10.1007/s10722-007-9260-3
Bory S, Lubinsky P, Risterucci A-M et al (2008c) Patterns of introduction and diversification of Vanilla planifolia (Orchidaceae) in Reunion Island (Indian Ocean). Am J Bot 95:805–815. https://doi.org/10.3732/ajb.2007332
Bouetard A, Lefeuvre P, Gigant R et al (2010) Evidence of transoceanic dispersion of the genus Vanilla based on plastid DNA phylogenetic analysis. Mol Phylogenet Evol 55:621–630. https://doi.org/10.1016/j.ympev.2010.01.021
Catchen J, Hohenlohe PA, Bassham S et al (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140. https://doi.org/10.1111/mec.12354
Chambers A (2021) Advancing vanilla genomics and plant breeding for the Americas. IV Congreso Internacional de Vainilla https://www.youtube.com/watch?v=vSElz3cYpJI, 5:47:00
Constantin D, Bois J (1915) Sur trois types de vanilles commerciales de TAHITI. Comptes Rendus De L’académie Des Sciences De Paris 161:196–202
Correll DS (1953) Vanilla-its botany, history, cultivation and economic import. Econ Bot 7:291–358. https://doi.org/10.1007/BF02930810
Danecek P, Auton A, Abecasis G et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Díaz-Bautista M, Francisco-Ambrosio G, Espinoza-Pérez J et al (2018) Morphological and phytochemical data of Vanilla species in Mexico. Data Brief 20:1730–1738. https://doi.org/10.1016/j.dib.2018.08.212
Earl DA, vonHoldt BM (2012) Structure harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conservation Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Elias M, Mühlen GS, McKey D et al (2004) Genetic diversity of traditional South American landraces of Cassava (Manihot Esculenta Crantz): an analysis using microsatellites. Econ Bot 58:242–256. https://doi.org/10.1663/0013-0001(2004)058[0242:GDOTSA]2.0.CO;2
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. https://doi.org/10.1371/journal.pone.0019379
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Favre F, Jourda C, Besse P, Charron C (2021) Genotyping-by-sequencing technology in plant taxonomy and phylogeny. In: Besse P (ed) Molecular plant taxonomy. Springer, New York, pp 167–178
Fischer MC, Rellstab C, Leuzinger M et al (2017) Estimating genomic diversity and population differentiation—an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genomics 18:69. https://doi.org/10.1186/s12864-016-3459-7
Gigant R, Bory S, Grisoni M, Besse P (2011) Biodiversity and Evolution in the Vanilla Genus. In: The dynamical processes of biodiversity - case studies of evolution and spatial distribution, InTech. pp 1–26
Gigant RL, De Bruyn A, M’sa T et al (2016) Combining pollination ecology and fine-scale spatial genetic structure analysis to unravel the reproductive strategy of an insular threatened orchid. S Afr J Bot 105:25–35. https://doi.org/10.1016/j.sajb.2016.02.205
Grisoni M, Dijoux J-B (2017) Vanilla variety named “Handa.” USA patent application US14/999,830
Hasing T, Tang H, Brym M et al (2020) A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nat Food 1:811–819. https://doi.org/10.1038/s43016-020-00197-2
Hu Y, Resende MFR, Bombarely A et al (2019) Genomics-based diversity analysis of Vanilla species using a Vanilla planifolia draft genome and genotyping-by-sequencing. Sci Rep 9:3416. https://doi.org/10.1038/s41598-019-40144-1
Jakše J, Kindlhofer K, Javornik B (2001) Assessment of genetic variation and differentiation of hop genotypes by microsatellite and AFLP markers. Génome 44:773–782. https://doi.org/10.1139/gen-44-5-773
Karremans AP, Chinchilla IF, Rojas-Alvarado G, et al (2020) A reappraisal of Neotropical Vanilla. With a note on taxonomic inflation and the importance of alpha taxonomy in biological studies. Lankesteriana. https://doi.org/10.15517/lank.v20i3.45203
Knaus BJ, Grünwald NJ (2017) vcfr : a package to manipulate and visualize variant call format data in R. Mol Ecol Resour 17:44–53. https://doi.org/10.1111/1755-0998.12549
Lepers-Andrzejewski S, Siljak-Yakovlev S, Brown SC et al (2011) Diversity and dynamics of plant genome size: an example of polysomaty from a cytogenetic study of Tahitian vanilla (Vanilla ×tahitensis, Orchidaceae). Am J Bot 98:986–997. https://doi.org/10.3732/ajb.1000415
Lepers-Andrzejewski S, Causse S, Caromel B et al (2012) Genetic linkage map and diversity analysis of Tahitian Vanilla ( Vanilla × tahitensis, Orchidaceae). Crop Sci 52:795–806. https://doi.org/10.2135/cropsci2010.11.0634
Lubinsky P, Cameron KM, Molina MC et al (2008) Neotropical roots of a Polynesian spice: the hybrid origin of Tahitian vanilla, Vanilla tahitensis (Orchidaceae). Am J Bot 95:1040–1047. https://doi.org/10.3732/ajb.0800067
Lubinsky P, Romero-González GA, Heredia SM, Zabel S (2010) Origins and patterns of Vanilla cultivation in tropical America (1500–1900): no support for an independent domestication of Vanilla in South America. In: Havkin-Frenkel D, Belanger FC (eds) Handbook of Vanilla science and technology. Wiley-Blackwell, Oxford, pp 117–138
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. https://doi.org/10.14806/ej.17.1.200
Martínez-Ainsworth NE, Tenaillon MI (2016) Superheroes and masterminds of plant domestication. CR Biol 339:268–273. https://doi.org/10.1016/j.crvi.2016.05.005
McKey D, Elias M, Pujol B, Duputié A (2010) The evolutionary ecology of clonally propagated domesticated plants: Tansley review. New Phytol 186:318–332. https://doi.org/10.1111/j.1469-8137.2010.03210.x
Merrill ED (1954) The botany of Cook’s Voyages and it’s Unexpected Significiance in Relation to Anthropology, Biogeography and History (Chronica Botanica). Waltham, Massachusetts
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Perrier X, Jacquemoud-Collet JP (2006) DARwin: dissimilarity analysis and representation for windows. https://darwin.cirad.fr/index.php
Piperidis N, D’Hont A (2020) Sugarcane genome architecture decrypted with chromosome-specific oligo probes. Plant J 103:2039–2051. https://doi.org/10.1111/tpj.14881
Portères R (1954) Le genre Vanilla et ses espèces. Le vanillier et la vanille dans le monde. Paris, Editions Paul Lechevalier XL VI:94–290
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pujol B, Mühlen G, Garwood N et al (2005) Evolution under domestication: contrasting functional morphology of seedlings in domesticated cassava and its closest wild relatives. New Phytol 166:305–318. https://doi.org/10.1111/j.1469-8137.2004.01295.x
R Development Core Team (2010) A language and environment for statistical computing: reference index. R Foundation for Statistical Computing, Vienna
Rambaut A (2006) FigTree. http://tree.bio.ed.ac.uk/software/figtree/
Roux-Cuvelier M, Grisoni M, Bellec A et al (2021) Conservation of horticultural genetic resources in France. Chronica Horticulturae 61:21–36
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. In: University of Kansas Science Bulletin. pp 1409–1438
Soto Arenas MÁ (1999) Filogeografía y recursos genéticos de las vainillas de México. Instituto Chinoin - Herbario de la Asociación Mexicana de Orquideología AC, Mexico
Soto Arenas MÁ (2003) Vanilla. In: Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) Genera orchidacearum: Orchidoideae. Owford University Press, USA, p 402
Soto Arenas MA, Cribb P (2013) A new infrageneric classification and synopsis of the genus Vanilla Plum. ex mill. (Orchidaceae: Vanillinae). Lankesteriana. https://doi.org/10.15517/lank.v0i0.12071
Watteyn C, Scaccabarozzi D, Muys B et al (2022) Trick or treat? Pollinator attraction in Vanilla pompona (Orchidaceae). Biotropica 54:268–274. https://doi.org/10.1111/btp.13034
Zohary D (2004) Unconscious selection and the evolution of domesticated plants. Econ Bot 58:5–10. https://doi.org/10.1663/0013-0001(2004)058[0005:USATEO]2.0.CO;2
Acknowledgements
We are grateful to Katia Jade for preparing tissue cultured plant material and to the Plant Protection Platform (3P, IBISA) for lab facilities and plant resources (BRC Vatel) access. We would like to thank the SouthGreen Bioinformatics Platform (http://www.southgreen.fr/) for computational resources and the GeT-PlaGe platform (INRAE, Toulouse, France) for Illumina sequencing. KeyGene N.V. owns patents and patent applications protecting its Sequence Based Genotyping technologies.
Funding
This research was funded by the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD) and the Université de la Réunion. Felicien Favre was supported by a MENRT grant from the French Ministry of Higher Education, Research and Innovation. This work was also funded by the European Regional Development Fund (ERDF), the Conseil Régional de la Réunion, and the Conseil Général de la Réunion. This work was carried out with the financial support of FORDECYT-CONACYT through the Vaniclim Project (Number 297484) “Estrategias para la adaptacion y mitigacion al cambio climatico necesarias para el rescate del cultivo de la vainilla en Mexico.
Author information
Authors and Affiliations
Contributions
CC, MG and CJ conceived and designed the experiment. JBD produced hybrids and grew the plant material at CRB Vatel. SL provided V. × tahitensis from the Etablissement Vanille de Tahiti (EVT) and provided her expertise on the genetics of V. × tahitensis. JH extracted DNA, RR produced GBS libraries, CJ performed the SNPs calling and QP the genomic mapping of SNPs. FF and CC analyzed the data and wrote the manuscript. PB, MG and CJ contributed to and edited the manuscript. All authors have reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Favre, F., Jourda, C., Grisoni, M. et al. A genome-wide assessment of the genetic diversity, evolution and relationships with allied species of the clonally propagated crop Vanilla planifolia Jacks. ex Andrews. Genet Resour Crop Evol 69, 2125–2139 (2022). https://doi.org/10.1007/s10722-022-01362-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01362-1