Abstract
Sustainable barley (Hordeum vulgare L.) production will require access to diverse ex-situ conserved collections to develop varieties with high yields and capable of overcoming the challenges imposed by major abiotic and biotic stresses. This study aimed at searching efficient approaches for the identification of new sources of resistance to barley leaf rust (Puccinia hordei Otth). Two subsets, Generation Challenge Program Reference set (GCP) with 188 accessions and leaf rust subset constructed using the filtering approach of the Focused Identification of Germplasm Strategy (FIGS) with 86 accessions, were evaluated for the seedling as well as the adult plant stage resistance (APR) using two barley leaf rust (LR) isolates (ISO-SAT and ISO-MRC) and in four environments in Morocco, respectively. Both subsets yielded a high percent of accessions with a moderately resistant (MR) reaction to the two LR isolates at the seedling stage. For APR, more than 50% of the accessions showed resistant reactions in SAT2018 and GCH2018, while this rate was less than 20% in SAT2017 and SAT2019. Statistical analysis using chi-square test of independence revealed the dependency of LR reaction on subsets at the seedling (ISO-MRC), as well as at the APR (SAT2017 and SAT2018) stage. At seedling stage, the test of goodness of fit showed that GCP subset yielded higher percentages of resistant accessions than FIGS-LR in case of ISO-MRC isolate but the two subsets did not differ for ISO-SAT. At APR, FIGS approach performed better than GCP in yielding higher percentages of accessions in case of SAT2017 and SAT2018. Although some of the tested machine learning models had moderate to high accuracies, none of them was able to find a strong and significant relationship between the reaction to LR and the environmental conditions showing the needs for more fine tuning of approaches for efficient mining of genetic resources using machine learning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cultivated barley (Hordeum vulgare subsp. vulgare L.) is the fourth most important cereal crop in the world after wheat, maize, and rice, in terms of production of 143.13 million metric tons and acreage of around 47.37 million hectares (FAOSTAT 2017). In Morocco, barley is grown on 2 million hectares in the arid and semi-arid regions with 1.23 t/ha of average grain yield, which is relatively low compared to North America (3.67 t/h) (FAOSTAT 2017). The lower national average grain yield of barley is due to limited or no use of inputs, and the prevalence of abiotic and biotic constraints. Foliar diseases such as powdery mildew (Blumeria graminis f. sp. hordei), net form net blotch (Pyrenophora teres f. teres) and spot form net blotch (Pyrenophora teres f. maculata), spot blotch (Cochliobolus sativus), and leaf rust (Puccinia hordei) are important biotic constraints that limit the grain and straw yields and their quality. Barley leaf rust caused by Puccinia hordei Otth (Ph) is one of the most destructive and globally spread barley diseases (Clifford 1985; Park et al. 2015). It is widely distributed throughout barley growing region, and can cause serious yield losses in the regions of North Africa, Europe, New Zealand, Australia, the Eastern and Midwestern parts of United States, and some parts of Asia, where susceptible and late maturing varieties of barley are sown (Arnst et al. 1979; Clifford 1985; Chicaiza et al. 1996; Brunner et al. 2000; Niks et al. 2000). Losses of barley production due to LR can reach up to 30% on susceptible cultivars (Cotterill et al. 1992; Griffey et al. 1994).
Applying fungicides is an efficient strategy to control major foliar diseases, but it is not economical for barley grown under marginal lands and low-input conditions of Morocco. Therefore, the use of resistant varieties is the most effective, economical, and environmentally safe way for controlling barley LR. This can be achieved by transferring identified resistant genes from diverse genetic resources into elite barley germplasm (Hajjar and Hodgkin, 2007; Rehman et al. 2020). To date, 23 Rph genes conferring hypersensitive resistance to barley leaf rust at the seedling stage (Rph1-Rph19, Rph21, Rph22, Rph25 and Rph28), and 3 APR genes (Rph20, Rph23, Rph24) have been identified from Hordeum vulgare subsp. vulgare, or transferred from H. vulgare subsp. spontaneum, and H. bulbosum (Park 2015; Kavanagh et al. 2017; Yu et al. 2018; Rothwell et al. 2020; Mehnaz et al. 2021). However, LR resistance faces a big challenge from a rapidly evolving pathogen due to recombination and mutations which lead to the development of new pathotypes that overcome deployed single major Rph genes in a short time span (McIntosh 1988; Figueroa et al. 2016). Most of the barley varieties released in Morocco are susceptible to LR, and limited sources of resistance are available against the LR populations prevailing in the northern parts of Morocco. Therefore, it is required to evaluate and identify continuously new sources of resistance from existing germplasm, and from gene bank collections (Qualset 1975; Sing et al. 2015).
The genetic resources remain the most important source of parental germplasm for barley breeding programs to develop new varieties with high yield, better end-use quality, tolerant to abiotic stresses, and resistant to major diseases and pests. But the search for a given trait is limited owing to the large number of accessions being held in the genebanks. Further, the evaluation of these large collections for some traits can be very expensive. To facilitate the screening and the mining of genetic resources, it requires the development of intelligent sub-setting approaches to fit the available funding and facilities (ICARDA 2015). These approaches aim to select subsets from the original collection to harness maximum diversity within limited number of accessions (Gollin et al. 2000a, b). Frankel and Brown (1984) recommended the use of core collection which selects 5–10% of the original collection, representing maximum geographical or morphological diversities. However, because of the large number of accessions in the entire collection in genebanks, even a core collection can still be unmanageable for the evaluation of some traits, and other sub-setting approaches were suggested. Mini-core collections were suggested by Upadhyaya and Ortiz (2001) to concentrate broad genetic diversity in smaller subsets. It allows selecting about 1% of the total accessions from the entire collection to represent maximum diversity. The Generation Challenge Program (GCP) (https://www.generationcp.org) recommended the development of a reference set representing 10% of the core collection to represent maximum diversity using molecular markers.
From ICARDA barley in-trust collection totaling more than 32,000 accessions, the barley core collection (composite set) of 3000 accessions of both cultivated and wild progenitor species (H. spontaneum) was selected based on climatological data of the collection sites; from which the Generation Challenge Program (GCP) developed a reference subset of 300 accessions based on the diversity of EST-derived, and genomic SSR markers (https://www.croptrust.org/wp/wp-content/uploads/2014/12/Barley_Strategy_FINAL_27Oct08.pdf), however, many researchers have reported on the limitations of core collections in capturing rare and adaptive alleles (Dwivedi et al. 2008; Xu 2010).
The Focused Identification of Germplasm Strategy (FIGS) was developed by ICARDA in collaboration with the Australian and the Russian partners as an alternative approach for efficient mining of genetic resources that maximize the likelihood of capturing specific adaptive traits in subsets of manageable size extracted from the original collection (Mackay 1990; Street et al. 2008). FIGS is based on the co-evolution between the accessions and the environmental conditions in which they evolved (Mackay 1995; Gollin et al. 2000a, b; Mackay and Street 2004; Bari et al. 2012). This approach exploits the development of the relationship between the specific sought-trait and ecogeographical data by filtering germplasm collections through exerting selection pressures of the emergence of a sought trait. When the relationship is confirmed, a manageable subset can be selected to include accessions with high probability of having the sought trait. FIGS subsets have allowed to identify for the first-time sources of resistance to Sunn pest in wheat (El Bouhssini et al. 2009), resistance to net blotch in barley (Endersen et al. 2011), and drought tolerance in faba bean (Khazaei et al. 2021).
The present study aimed at: (i) identification of sources of resistance to LR in FIGS_LR and GCP subsets; (ii) assessing the dependence of resistance on sub-setting approach; and (iii) search for the best model that describes the relationship between resistance to LR, and the environmental conditions using machine learning.
Materials and methods
Plant material
Two barley subsets extracted from ICARDA in-trust collection available from the regeneration efforts conducted in Morocco to reconstruct the active and base collections were used in this study. A total of 188 accessions from the reference set constructed within the Generation Challenge Program (GCP) and extracted from the composite set of barley collection held at ICARDA based on diversity of EST-derived and genomic SSR markers (Supplementary Table S1). Another subset composed of 86 accessions was selected using filtering approach of the Focused Identification of Germplasm Strategy (FIGS_LR) based on the following parameters:
-
Count number of days where the average daily temperature is between 8–15 °C, 10 days before the onset of growing period and up to 15% into the vegetative phase.
-
Remove sites with zero count from step 1.
-
Sum daily rain for 10 days before the onset of growing period up to 10% into the vegetative phase.
-
Normalize both variables (steps 1 and 2) to range 0–1 for each site.
-
Add variables to create index 1.
-
Rank based on index 1 and remove bottom 25 percent of sites.
For the remaining sites, the following was done:
-
From 10% into the vegetative phase until onset of grain filling divide into 3 separate sub-phases of equal length.
-
For each sub-phase count the number of days where the average daily temperature is between 18–20 oC.
-
For each sub-phase determine the amount of precipitation.
-
Remove sites if any of the variables = 0 (3 count variables and 3 precipitation variables).
-
Normalize each variable for a range between 0–1.
-
Add each variable and then add index 1 to create index 2.
-
Rank sites using index 2 from largest to smallest.
-
Since there are more sites than the desired set size, then one accession could be chosen randomly from each site starting at the top ranked site until the desired set size is reached. Alternatively, this approach could be taken after one candidate accession is donated by each country represented in the candidate site list.
The climatic conditions layers were extracted from the GIS surfaces modeled from data collection sites as described by De Pauw (2008). The FIGS_LR subset has more accessions from Greece, Turkey, Ethiopia, and India (Supplementary Table S2).
Seedling screening of LR resistance
The seedling screening of GCP and FIGS_LR subsets was conducted under controlled conditions in the growth chamber with two pure isolates of LR (ISO-SAT and ISO-MRC). The single urediniospore was isolated from infected leaves collected from the experimental stations of Sidi Allal Tazi (ISO-SAT) and Marchouch (ISO-MRC) in 2017 and were multiplied on the susceptible barley cultivars (Bowman and Aglou) followed by collection and drying of urediniospores on silica gel and storage at − 80 ºC until further use.
Barley plants were grown in sterilized peat moss (supplemented with 14–14–14 NPK) in plastic cones (14 cm long cones with 3.8 cm diameter) positioned in a 14 × 7-unit tray (Steuwe & Sons, Inc., OR, United States). For each barley accession, 4–5 seeds were planted per cone in two replications. Each tray contained 96-test genotypes along with resistant (Philadelphia) and susceptible (Bowman) checks. Plants were raised in the growth chamber (Snijder Scientific, Tilburg, the Netherlands) with a photoperiod of 16 h light/8 h dark at 20 ± 1 ºC. Inoculation was carried out on 10–12 days old seedlings when the first leaf was fully expanded. To prepare LR inoculum, urediospores were taken from the − 80 °C freezer and subjected to heat shock for 5 min at 40 °C.
For each tray, 15 mg of urediniospores were suspended in 10 ml of light mineral oil (Novec 7100, Sigma Aldrich), and this spore suspension was sprayed onto plants as a fine mist using an airbrush (Revell, Munchen, Germany). Inoculated plants were left to dry for 20 min at the room temperature and were placed in growth chamber in the dark for 24 h at 18 °C with ~ 100% relative humidity. Then plants were maintained in the growth chamber with a light/ dark period of 16/8 h at 20 °C for symptoms development. The evaluation for LR reaction was carried out 12–14 days post-inoculation based on infection types (ITs) according to the 0 to 4 scale developed by Stakman et al. (1962). The seedlings were classified either as immune (0), resistant (0; and 1), moderately resistant (2), moderately susceptible (3), or susceptible (4).
Field screening of LR resistance
The field screening of LR resistance was performed at the INRA experimental station of Sidi Allal Tazi (34° 52' N, 6.32 W) during 2016–17 (SAT2017), 2017–18 (SAT2018), 2018–19 (SAT2019), at Marchouch (33° 56′10″N 6°69′21″W) during 2017–18 (MRC2018), and at Guich (33°58′59.7″N 6°51′41.6″W) during 2017–18 (GCH18). The tested accessions were sown as single rows of 1 m with 0.5 m row spacing between adjacent accessions using an augmented block design. The seed mixture of susceptible cultivars Bowman and Aglou were sown as a rust spreader row at the border of each block to allow the uniform distribution of LR inoculum.
At SAT, the disease was established naturally, but at Guich station the disease was initiated using the artificial inoculation. About 1 g of dried urediniospores were suspended in 200 ml of mineral oil and sprayed on the trial using an airbrush (Revell, Munchen, Germany). The establishment and spread of the disease were favored by covering the spreader rows with a plastic sheet overnight and by periodic sprinkler irrigation. The LR resistance was assessed for GCP and FIGS_LR subsets at growth stage 65–77 (Zadoks et al. 1974) using the modified Cobb scale (Peterson et al. 1948) which combined the LR severity (0 to 100%) and host response; 0 (Immune), no visible infection on plants; R (resistant), visible chlorosis or necrosis, no uredia are present; MR (moderately resistant), small uredia are present and surrounded by either chlorotic or necrotic areas; MS (moderately susceptible), medium sized uredia are present and possibly surrounded by chlorotic areas; S (susceptible), large uredia are present, generally with little or no chlorosis and no necrosis. The Coefficient of Infection (CI) was calculated by multiplying the infection response values (R = 0.2, MR = 0.4, MS = 0.8, S = 1) with the percent disease severity (0–100%) (Stubbs et al.1986), and the accessions were rated based on the average coefficient of infection (ACI) where values of 0–7, 8–16, 17–29, 30–50, and > 50 were considered as resistant, moderately resistant, moderately susceptible, susceptible, and highly susceptible, respectively.
Data analysis
Comparing the reactions of GCP and FIGS subsets
The statistical analysis was performed using R software (R Core Team 2018). The statistical association between sub-setting approach and the reaction to LR was calculated using χ2 test of independence with significance level (α = 0.05) using the following equation:
The equation used for calculating expected values in a test of independence was as follows:
where \({E}_{ij}\) = expected value, \(\sum_{k=1}^{c}{O}_{ij}\) is the sum of the ith column, \(\sum_{k=1}^{r}{O}_{kj}\) is the sum of the kth row, N is the total number.
To find out the differences between FIGS and GCP subsets in terms of reaction to LR, the test of goodness of fit using χ2 test at a significance level (α = 0.05) was used where GCP was simulated to a random sample. The expected values for the test of goodness of fit are calculated as follows:
where Ei is the expected value, n is the total sample size, and pi is the hypothesized proportion of observations in level i.
Both tests were performed using different groupings of reactions, all classes (I, R, MR, MS and S), three classes (I + R, MR + MS, S) and (I + R + MR, MS, S), two classes (I + R + MR, MS + S) at the seedling stage, and all classes (R, MR, MS, S, HS), three classes (R, MR + MS, and S + HS), and two classes (R + MR + MS, S + HS) at the adult plant stage.
Modeling of the reaction to leaf rust disease
The second pathway of FIGS using machine learning was investigated using the available reactions of the accessions of FIGS_LR and GCP subsets to find a function that links adaptive traits, environments (and associated selection pressures) with genebank accessions. We used environmental data from WorldClim1 databases as predictors. The WorldClim is an open access database providing global climatic layers describing past climatic profiles of collection sites intended for spatial modeling or mapping. It includes averages of monthly minimum and maximum temperatures, precipitation and bioclimatic variables (Fick and Hijmans 2017).
The following machine learning algorithms were used: K-nearest neighbors KNN (Kotsiantis 2007), Support Vector Machine SVM (Hsu et al. 2010), Random Forest RF (Breiman, 2001), Neural networks NNET (Venables and Ripley 2002), and Bagged Carts BCART (Kołcz 2000). Each machine learning model was tuned to select the best tuning parameters using a training set (70% of the total set), and then the best model was selected between different machine learning models based on several metrics including accuracy, specificity, and Kappa. The modeling metrics were computed on the test set (30% of the total set). In this study, R language and caret library were used for machine learning analysis (Kuhn 2008). Models were tuned for parameter’s optimization and trained on 70% of the data and tested with 10 cross validation folds and 100 replications. In addition, modeling was done for the two isolates for the seedling stage. For the APR, modeling was done for the entire multi-locations data sets and for each location separately.
Results
Seedling resistance
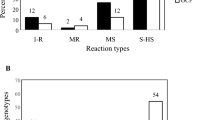
In the seedling test, successful artificial inoculation was carried out for the two isolates and diverse infection responses were recorded. The frequency distribution of infection response of GCP and FIGS_LR accessions at the seedling stage has been presented in Fig. 1. The uniformity of the disease development was assessed through the high susceptibility of the check Bowman over the two replications. The highest number of accessions of both FIGS_LR (65.88% for ISO-MRC, and 51.16% for ISO-SAT), and GCP (62.87 and 48.4% for ISO-MRC and ISO-SAT isolates, respectively) subsets showed MR reaction, and only few accessions showed R reaction with one accession of GCP having immune reaction (Figs. 1a and b). More resistant accessions were noted when tested to ISO-MRC isolate with 5.88% for FIGS_LR and 22.75% for GCP compared with ISO-SAT isolate that showed 10.47% and 13.3% resistant accessions for FIGS_LR and GCP, respectively. Some accessions showed different disease reaction for the two isolates. For example, the accessions IG 143,872, IG 143,862, IG 143,864, IG 143,872, IG 143,890, IG 143,978, IG 143,984, IG 144,006, IG 144,090, IG 144,029, IG 144,077, IG 143,871, and IG 144,012 were resistant to ISO-MRC isolate but not to ISO-AT isolate, whereas accessions IG 143,963, IG 18,725, IG 19,525 were resistant to ISO-AT isolate but not to ISO-MRC isolate. Ten barley accessions in GCP (IG 143,876, IG 143,886, IG 143,906, IG 143,929, IG 143,998, IG 143,999, IG 144,014, IG 144,064, IG 144,076, IG 144,108) and one in FIGS_LR (IG 18,957) were resistant to both isolates. Most of the resistant accessions originated from USA, Turkey, Greece, and Morocco.
Adult plant resistance (APR)
Under field conditions, good natural LR infection was recorded at the Sidi Allal Tazi during the three cropping seasons, and good artificial infection was established at Guich in 2018. However, late and light artificial infection at Marchouch during 2017 and 2018 seasons did not allow disease severity assessments. The uniformity of the disease development was assessed through the high susceptibility of the checks, Bowman and Aglou, at the adult stage at Sidi Allal Tazi and Guich sites. The good development of the LR allowed efficient screening of the germplasm at the adult plant stage, as shown by wide range of reactions observed (Fig. 2). The average of coefficient of infection (ACI) values across the environments ranged from 0 to 85 with several accessions showing contrasting reactions in different environments. Accessions of FIGS_LR and GCP subsets showed different distributions of the reaction classes with near normal distribution for SAT2017 and SAT2019, and positive skewness with high percentage of R accessions (ranged from 55.8 to 68.3%) for SAT2018 and GCH2018 (Fig. 2b and c). While at SAT2017 and SAT2019, this reaction class distribution percentage ranged from 5.59 to 19.64% (Fig.. 2a and d) for both subsets. When considering MR reaction, additional 8.94 to 30.36% accessions were found during SAT2017 and SAT2019 cropping seasons. The highest numbers of susceptible accessions were observed at SAT2017 with 34.64 and 15.29% showing susceptibility, and 22.91% and 15.29% were highly susceptible (HS) for GCP and FIGS_LR, respectively. Only three accessions in FIGS_LR (IG 28,636, IG 28,647 and IG 33,039), and four accessions in GCP subset (IG 143,945, IG 144,000, IG 144,064, IG 144,105) were found to be resistant across all environments (Table 1).
Frequency distributions of the adult plant resistance (APR) of FIGS_LR and GCP subsets to leaf rust under field conditions at Sidi Allal Tazi during a 2017 (SAT2017), b 2018 (SAT2018), d 2019 (SAT2019) cropping seasons, and at Guich during 2018 (GCH2018 (c)). Here R = resistant, MR = moderately resistant, MS = moderately susceptible, and S = susceptible
Comparison of the reaction of FIGS_LR and GCP to leaf rust
Applying the chi-square tests of independence to both subsets, the results showed that there is a significant relationship (P-value = 0.04) between the response to LR disease and the sub-setting approach for ISO-MRC isolate, but not for ISO-SAT, when considering all classes of reaction. For ISO-MRC, GCP included one accession with immune reaction and 23% of the accessions being resistant, while FIGS_LR subset showed only 6% of the accessions being resistant (Table 2, Fig. 1a). When screened with ISO-SAT isolate, both FIGS_LR and GCP showed that the infection response to LR was not dependent on the sub-setting (p = 0.85), displaying same distribution patterns of the reaction. No accession was found immune in both subsets while 9 (10.47%) and 25 (13.3%) accessions were resistant, and 44 (51.16%) and 91 (48.4%) accessions were MR for FIGS_LR and GCP subsets, respectively (Table 2, Fig. 1b).
The test of goodness of fit showed that GCP subset yielded higher percentage of accessions with R, but FIGS_LR subset yielded higher percentage of accessions with MR reactions in case of ISO-MRC isolate, but no significant differences were observed between the two subsets when tested with ISO-SAT isolate (Table 2).
Except the grouping of two classes (I + R + MR, MS + S) for the goodness of fit test, the tests of independence and goodness of fit were significant for ISO-MRC, but not for ISO-SAT for different groupings of the reactions (Table 3).
For APR, the reaction to LR was dependent on subsets for Sidi Allal Tazi during 2017 (SAT2017) and 2018 (SAT2018) with respective χ2 (P-values) of 0.001 and 0.02, respectively. But this dependence was not found in case of GCH 2018 (GCH18) and Sidi Allal Tazi 2019 (SAT2019) (Table 4). The tests of goodness of fit showed that FIGS_LR outperformed GCP subset at Sidi Allal Tazi with higher percentages of accessions with R and MR reactions under heavy infection in 2017, but the opposite was observed during 2018 season at the same site. For GCH2018 and SAT2019 environments, no significant differences were observed between the two subsets.
When different groupings of the reactions were performed, the significance probability of the two tests were highly significant for SAT2017 and SAT2018, but not for GCH2018 and SAT2019, except for the test of goodness of fit for GCH2018 in case of the grouping (R + MR + MS; S + HS) with P-value of 0.04 (Table 5).
Predictive modeling of the reaction to leaf rust
At the seedling stage, the tested machine learning models did not perform similarly for the two LR isolates. For ISO-MRC isolate, all models yielded a significant medium to very high accuracy. The maximum accuracy (0.94) was reached using the BCART model and was then chosen as the best model. The remaining modeling parameters showed the strong mathematical relationship between the reaction to ISO-MRC and the environmental characteristics (Table 6). However, the modeling pattern was opposite for ISO-SAT isolate where all the models were not significantly accurate (Table 7), since the accuracy was similar to the “No Information Rate” and hence demonstrating that the models were as good as the naïve model. It is noticeable that the specificity was much lower than sensitivity for all tested models.
For the APR, no model performed significantly for the two locations (Table 8). Accuracy was high for all models, however, the unbalanced data due to the higher number of resistant genotypes make the model not performing better than the naïve model because of the low values of specificity and high value of “No Information Rate”. Among the tested models, RF was the best model for all locations.
Discussion
LR occurs annually with high incidence in the Northern regions of Morocco, and the Sidi Allal Tazi has been used as the LR hotspot for the barley germplasm screening. Over three years, most of the barley varieties and advanced breeding lines showed high susceptibility to P. hordei at this site. The development of high yielding varieties with adequate levels of resistance is the key to an integrated management of LR and is compatible with the general consideration of barley as a low input crop by most farmers. Recent studies on the screening of international germplasm collections reported that the effective sources of LR resistance available are limited, highlighting the needs for additional new sources of resistance (Golegaonkar et al. 2009; Derevnina et al. 2013; Sandhu et al. 2014; Singh et al. 2015). Genetic resources conserved ex situ in the genebanks are important sources of breeders ‘sought traits including the resistance to major diseases, but need efficient mining approaches. In this study, both FIGS_LR and GCP subsets have yielded sources of resistance to LR, but only few accessions showed resistant reaction at the seedling and/or adult plant stages in SAT2017 and SAT2019. The results indicated the changes in the reaction of accessions to different isolates at the seedling stage and at different locations and in different years at the adult plant stage.
The response of differentials to pure isolates of a pathogen at SRT and to field populations at APR decipher putative R genes effective at both stages. Race analysis of both isolates of LR (ISO-MRC and ISO-SAT) on 19-Bowman near isogenic lines (NILs) revealed their diverse virulence spectrum (Amouzoune et al. unpublished data). ISO-MRC was virulent on NILs carrying Rph2.b, Rph3.c, Rph4.d, Rph5.e, Rph6.f Rph5, Rph7.g, Rph8.h, Rph9.i, Rph10.o, Rph11.p, Rph9.z Rph12, Rph2.j, Rph2.y, Rph2.t, whereas the isolate ISO-SAT was virulent on NILs carrying Rph1.a, Rph3.c, Rph4.d, Rph8.h, Rph9. Furthermore, of the 19 differentials tested, 11 (58%) showed differential interaction between both isolates. Hence, differential response of both LR isolates to FIGS_LR and GCP at SRT can be attributed to their diverse virulence spectrum (Fig. 1). In addition, among the resistant accessions to both isolates in case of FIGS_LR and GCP subsets, only 7 and 16% R accessions were common which further corroborate difference in their virulence spectrum. Of the 19 Bowman differential lines tested at the adult plant stage at Sidi Allal Tazi in 2017 cropping season (SAT2017), only one differential line carrying Rph2 (Rph2.y) displayed moderately resistant reaction to LR field population (unpublished data). Contrary to SRT, FIGS_LR performed better than GCP at APR. Except SAT2018, higher percentage of R and MR barley accessions were observed in SAT2017, SAT2019, and GCH2018 in FIGS subset compared to GCP (Fig. 2). In the present study, four accessions IG 143945, IG 144000, IG 144064 from GCP subset, and three accessions IG 28613, IG 28636, IG 33039 from FIGS_LR subset showed resistant (R) to moderately resistant (MR) response at the seedling and at APR stage (Table 1). Under Moroccan growing conditions, barley is planted in November and LR is the last disease which effect barley in March–April. Therefore, APR is quite important, and a large number of R-MR accessions identified in FIGS_LR and GCP subsets will be useful resource for combating LR. Most probably, LR resistant accessions identified in this study may possess either new R genes or allelic variants of existing R genes or a combination of both. A high-density genotyping and genome wide association studies seem to be a logical step to dissect the resistance diversity. These putative R genes could be either pyramided or used sequentially to ensure a better R gene deployment strategy.
The seedling resistance is usually characterized by hypersensitivity and is governed by single major genes, Such genes can be easily overcome by new LR races because of their excessive utilization over large areas which exert selection pressure on the pathogen population which lead to the emergence of new races, and eventual breakdown the effectiveness of resistance genes. Virulence has been detected for most known seedling Rph genes in various barley growing regions throughout the world. In Australia, only Rph3, Rph7, Rph11, Rph14, Rph15, and Rph18 of the characterized major genes were still effective to prevailing pathotypes (Cotterill et al. 1995; Park 2003). However, pathotypes virulent to Rph3 were detected in New Zealand (Cromey and Viljanen-Rollinson 1995), and the virulence for Rph7 has been identified in Israel (Golan et al.1978), Morocco (Parlevliet et al. 1981), and North America (Steffenson et al. 1993). Virulence for Rph11 and Rph14 has also been found frequently in many parts of the world (Fetch et al. 1998), and virulence to Rph15 was reported by Sun et al. (Sun 2007). Therefore, an accession with LR resistance at the seedling stage alone might not provide durable and effective resistance (Singh 1992; Park 2008; Singh et al. 2015).
APR against rusts is a key component of durable resistance in wheat (Singh et al., 2001). Similarly, APR to barley LR is a good strategy for effective disease control and the identification and characterization of such sources could facilitate their utilization in breeding programs. Since there are several accessions at the adult plant stage with MR and MS reactions or with slow progression of the disease based on the area under the disease progress curve (data not presented) under heavy rust epidemics, partial resistance and slow rusting mechanism could be considered to ensure a race non-specific and a more durable resistance. Several studies have promoted partial and non-race specific resistance in case of rusts and powdery mildew in barley and wheat as this type of resistance is available in some commercial varieties (Parlevliet and Kuiper 1977; Andres and Wilcoxon 1986; Niks et al. 2000; Stuthman et al. 2007). Several APR genes were well characterized and deployed in wheat to control rust diseases (Park and McIntosh 1994). In barley, three genes governing APR to LR have been identified and used (Rph20, Rph23, and Rph24) (Hickey et al. 2011; Singh et al. 2015; Ziems et al. 2017). Even if there are no reports of virulence for Rph20, Rph23 or Rph24, identifying new APR resistance genes for LR are essential for diversifying resistance and to promote gene pyramiding to increase resistance levels. Marker assisted selection (MAS) provides an opportunity to breeders to pyramid the APR genes in barley.
FIGS has shown its efficiency in identifying novel sources of resistance to powdery mildew, yellow and stem rusts, Sunn pest, and Russian wheat aphid in wheat (Bhullar et al. 2009; El Bouhssini et al. 2009, 2011; Bari et al. 2012, 2014), and to net blotch of barley (Endresen et al. 2011). This study included the first attempt to compare FIGS with another subset, the Reference set of the Generation Challenge Program (GCP) selected from the global barley core collection based on diversity using SSR markers. FIGS sub-setting using filtering approach has allowed to identify higher percentages of accessions when combining R and MR reactions compared to GCP subset in case of field tests (except SAT2018). The reduced sample size as well as the non-balance between the two classes (Resistant and Susceptible) could explain the low predictability of the machine learning models. Modeling outcomes using machine learning approach were dependent on the isolates or predominant field pathogen populations and the environments. The results showed the need for further fine tuning of FIGS approach to consider the diversity of virulence of the pathogen populations using larger subsets. Overall FIGS remains more relevant as it focuses on the traits needed by users, uses available evaluation data, and allows to select subsets from all the collections compared to core and mini-core collections where the focus is only on the overall genetic diversity included in 10% and 1% of the whole collection. It will be interesting also to compare both sub-setting methods in yielding new different effective genes. This can be investigated using molecular markers or by screening the identified sources of resistance to a larger number of isolates with different virulence spectrums.
Conclusion
This current study suggests that the trait mining approach can be an efficient alternative to the core collection method. The resistant and moderately resistant accessions at the seedling and at the adult plant stages in this study are valuable resources of P. hordei resistance and can lead towards effective and durable resistance against P. hordei when combined with appropriate gene deployment strategies. The evaluation of larger subsamples in different environments, and against different pathotypes will allow the fine tuning of FIGS sub-setting approach using machine leaning.
Data availability
The data that support the findings of this study are available in the ICARDA genebank database and can be obtained upon request from the corresponding author.
References
Andres MW, Wilcoxson RD (1986) Selection of barley for slow rusting resistance to leaf rust in epidemics of different severity. Crop Sci 26:511–514. https://doi.org/10.2135/cropsci1986.0011183X002600030015x
Arnst BJ, Martens JW, Wright GM, Burnett PA, Sanderson FR (1979) Incidence, importance and virulence of Puccinia hordei on barley in New Zealand. Ann Appl Biol 92:185–190
Bari A, Street K, Mackay M, Endresen DTF, De Pauw E (2012) Focused identification of germplasm strategy (FIGS) detects wheat stem rust resistance linked to environmental variables. Genet Resour Crop Evol 59:1465–1481
Bari A, Amri A, Street K, Mackay M, De Pauw E, Sanders R, Nazari K, Humeid B, Konopka J, Alo F (2014) Predicting resistance to stripe (yellow) rust (Puccinia striiformis) in wheat genetic resources using focused identification of germplasm strategy. J Agric Sci 152(06):906–916. https://doi.org/10.1017/S0021859613000543
Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B (2009) Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci USA 106:9519–9524
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Brunner S, Keller B, Feuillet C (2000) Molecular mapping of the Rph7.g leaf rust resistance gene in barley (Hordeum vulgare L.). Theor Appl Genet 101:783–788
Burdon JJ, BarrettLG RG, Thrall PH (2014) Guiding deployment of resistance in cereals using evolutionary principles. Evol Appl 7:609–624. https://doi.org/10.1111/eva.12175
Chicaiza O, Franckowiak JD, and Steffenson BJ (1996) New sources of resistance to leaf rust in barley. In: Slinkard AE, Scoles GJ, Rossnagel BG (eds) Proceedings of Fifth International Oat Conference & Seventh International Barley Genetics Symposium. Saskatoon, pp 706–708.
CGIAR Generation Challenge Programme (2012) Paper No 3: Genetic stocks. Position Papers: GCP’s research component. Texcoco, Mexico. pp 22
Clifford BC (1985) Barley leaf rust. In: Roelfs AP, Bushnell WR (eds) The cereal Rusts. Diseases, Distribution, Epidemiology and Control, Vol II. Harcourt Brace Jovanovich, Publishers Orlando, Florida 32887, pp 173–205
Cotterill PJ, Rees RG, Platz GJ, Dill-Macky R (1992) Effects of leaf rust on selected Australian barleys. Aus J Exp Agric 32:747–751
Cotterill PJ, Park RF, Rees RG (1995) Pathogenic specialization of Puccinia hordei Otth. in Australia, 1966–1990. Aus J Agric Res 46:127–134
Cromey MG, Viljanen-Rollinson SLH (1995) Virulence of Puccinia hordei on barley in New Zealand from 1990 to 1993. N Z J Crop Hortic Sci 23:115–119
Derevnina L, Singh D, Park RF (2013) Identification and characterization of seedling and adult plant resistance to Puccinia hordei in Chinese barley germplasm. Plant Breed 132(6):571–579. https://doi.org/10.1111/pbr.12082
Pauw De, Oweis T, Youssef J (2008) Integrating expert knowledge in GIS to locate biophysical potential for water harvesting: Methodology and a case study for Syria. ICARDA, Aleppo, Syria, p 59
Dwivedi SL, Stalker HT, Blair MW, Bertioli D, Upadhyaya HD, Nielen S et al (2008) Enhancing crop gene pool with beneficial traits using wild relatives. Plant Breed 30:179–230. https://doi.org/10.1002/9780470380130
El Bouhssini M, Street K, Joubi A, Ibrahim Z, Rihawi F (2009) Sources of wheat resistance to sunn pest, Eurygaster integriceps Puton, in Syria. Genet Resour Crop Evol 56:1065–1069
El Bouhssini M, Street K, Amri A, Mackay M, Ogbonnaya FC et al (2011) Sources of resistance in bread wheat to Russian wheat aphid (Diuraphis noxia) in Syria identified using the Focused Identification of Germplasm Strategy (FIGS). Plant Breed 130:96–97
Elmansour H, Singh D, Dracatos PM, Park RF (2017) Identification and characterization of seedling and adult plant resistance to Puccinia hordei in selected African barley germplasm. Springer, Dordrecht. https://doi.org/10.1007/s10681-017-1902-8
Endresen DTF, Street K, Mackay M, Bari A, De Pauw E (2011) Predictive association between biotic stress traits and ecogeographic data for wheat and barley landraces. Crop Sci 51:2036–2055
Endresen DTF, Street K, Mackay M, Bari A, Amri A, De Pauw E, Nazari K, Yahyaoui A (2012) Sources of resistance to stem rust (ug99) in bread wheat and durum wheat identified using focused identification of germplasm strategy (FIGS). Crop Sci 52:764–773
FAO. FAOStat, http://www.fao.org/faostat/en/#rankings/countries_by_commodity (2017)
Fetch TG, Steffenson BJ, Jin Y (1998) Worldwide virulence of Puccinia hordei on barley. Phytopathol 88:S28
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1km spatial resolution climate surfaces for global land areas. Int J Climatol 37(12):4302–4315
Figueroa M, Upadhyaya NM, Sperschneider J, Park RF, Szabo LJ, Steffenson B, Dodds PN (2016) Changing the game: using integrative genomics to probe virulence mechanisms of the stem rust pathogen Puccinia graminis f. sp. tritici. Front Plant Sci. https://doi.org/10.3389/fpls.2016.00205
Frankel OH, Brown AHD (1984) Current plant genetic resources- a critical appraisal. In: Chopra VL, Joshi BC, Sharma RP, Bansal HC (eds) Genetics: New Frontiers. Oxford & IBH Publ Co, New Delhi, India, pp 1–13
Global Crop Diversity Trust (2008) Global strategy for the ex situ conservation and use of barley germplasm [online]. Available from http://www.croptrust.org/documents/web/ BarleyStrategy_FINAL_27Oct08.pdf
Golan T, Anikster Y, Moseman JG, Wahl I (1978) A new virulent strain of Puccinia hordei. Euphytica 27:185–189
Golegaonkar PG, Singh D, Park RF (2009) Evaluation of seedling and adult plant resistance to Puccinia hordei in barley. Euphytica 166:183–197
Gollin D, Smale M, Skovmand B (2000a) Searching an ex situ collection of wheat genetic resources. Am J Agric Econ 82:812–827
Gollin D, Smale M, Skovmand B (2000b) Searching an ex situ collection of wheat genetic resources. Am J Agr Econ 82(4):812–827. https://doi.org/10.1111/0002-9092.00083
Griffey CA, Das MK, Baldwin RE, Waldenmaier CM (1994) Yield losses in winter barley resulting from a new race of Puccinia hordei in North America. Plant Dis 78:256–260
Hajjar R, Hodgkin T (2007) The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 156:1–13. https://doi.org/10.1007/s10681-007-9363-0
Hickey LT, Lawson W, Platz GJ, Dieters M, German S et al (2011) Mapping Rph20: a gene conferring adult plant resistance to Puccinia hordei in barley. Theor Appl Genet 123:44–68
Th H, van, F Menting, (2003) Diversity in ex situ genebank collections of barley. In: von Bothmer R, Th van H, Knüpffer H and Sato K, (eds) diversity in Barley (Hordeum vulgare). Elsevier Science BV, Amsterdam, The Netherlandspp, pp 247–257
Hsu CW, Chang CC, Lin CJ (2010) A practical guide to support vector classification. National Taiwan University, Taipei, Taiwan, Department of Computer Science
ICARDA (2013) A new approach to mining Agricultural gene banks – to Speed the pace of research Innovation for food security. “FIGS”–the Focused Identification of Germplasm Strategy. Research in Action 3. International Center for Agriculture Research in Dry Areas, Beirut, Lebanon.
ICARDA (2015) ICARDA Annual Report 2014. International Center for Agricultural Research in the Dry Areas, Beirut, Lebanon. 56 pp. ISSN: 92–9127–290–6
Kavanagh PJ, Singh D, Bansal UK, Park RF (2017) Inheritance and characterization of the new and rare gene Rph25 conferring seedling resistance in Hordeum vulgare against Puccinia hordei. Plant Breed 136:908–912
Khazaei H, Street K, Bari A, Mackay M, Stoddard FL (2013) The FIGS (Focused Identification of Germplasm Strategy) Approach Identifies Traits Related to Drought Adaptation in Vicia faba Genetic Resources. PLoS ONE 8(5):e63107. https://doi.org/10.1371/journal.pone.0063107
Khazaei H, O'Sullivan DM, Stoddard FL, Adhikari KN, Paull JG, Schulman AH, Andersen SU, Vandenberg A (2021) Recent advances in faba bean genetic and genomic tools for crop improvement. Legume Sci 3(3):e75. https://doi.org/10.1002/leg3.75
Knüpffer H, Hintum Th van (1995) The barley core collection: an international effort. In: (T. Hodgkin T, Brown AHD, Hintum Th van and Morales EAV (eds) Core Collections of Plant Genetic Resources John Wiley and Sons, Chichester, UK. pp. 171–178
Kołcz A (2000) N-tuple Network, CART, and Bagging, in Neural Computation,
Kotsiantis SB (2007) Supervised machine learning: a review of classification techniques, Informatica
Kuhn M (2008) Building Predictive Models in R Using the caret Package. J Stat Softw 28:1–26
Li LG, Cai L, Zhang XX, Zhang T (2014) Potentially novel copper resistance genes in copper-enriched activated sludge revealed by metagenomic analysis. Appl Microbiol Biotechnol 98:10255–10266
Mackay M (1990) Strategic planning for effective evaluation of plant germplasm. In: Srivastava JP, Damania AB (eds) Wheat genetic resources: meeting diverse needs. Wiley, Chichester, pp 21–25
Mackay M (1995) One core collection or many? In: Hodgkin T, Brown AHD, van Hintum TJL, Morales EAV (eds) Core collections of plant genetic resources. John Wiley & Sons Ltd, Chichester, pp 199–210
Mackay M, Street K (2004) Focused identification of germplasm strategy – FIGS. In: Black CK, Panozzo JF, Rebetzke GJ (eds) Proceedings of the 54th Australian Cereal Chemistry Conference and the 11th Wheat Breeders’ Assembly, pp 138–141.
McIntosh RA (1988) Catalogue of gene symbols for wheat. In: Miller TE, Koebner RMD (eds) Proceedings of the 7th International Wheat Genetics Symposium. Cambridge University Press, Cambridge, UK, pp 1225–1324
McIntosh MR, Royal Australian Chemical Institute (RACI) (1988) The Role of Specific Genes in Breeding for Durable Stem Rust Resistance in Wheat and Triticale. In: Simmonds S, Rajaram NW (eds) Breeding strategies for resistance to rust of wheat. CIMMYT, Mexico, DF, pp 1–9
Mehnaz M, Dracatos P, Pham A, March T, Maurer A, Pillen K, Forrest K, Kulkarni T, Pourkheirandish M, Park RF, Singh D (2021) Discovery and fine mapping of Rph28: a new gene conferring resistance to Puccinia hordei from wild barley. Theor Appl Genet. https://doi.org/10.1007/s00122-021-03814-1
Niks RE, Walther U, Jaiser H, Martínez F, Rubiales D, Andersen O, Flath K, Gymer P, Heinrichs F, Jonsson R, Kuntze L, Rasmussen M, Richter E (2000) Resistance against barley leaf rust (Puccinia hordei) in West-European spring barley germplasm. Ann appL Bid 92:185–190
Niks RE, Qi XQ, Marcel TC (2015) Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions and mechanisms. In: van Alfen NK (ed) Annual Review of Phytopathology pp 445–470
Park RF, McIntosh RA (1994) Adult plant resistances to Puccinia recondita f. sp. tritici in wheat. N Z J Crop Hortic Sci 22:151–158
Park RF (2003) Pathogenic specialization and pathotype distribution of Puccinia hordei in Australia, 1992 to 2001. Plant Dis 87:1311–1316
Park RF (2008) Breeding cereals for rust resistance in Australia. Plant Pathol 57:591–602. https://doi.org/10.1111/j.1365-3059.2008.01836.x
Park RF, Golegaonkar P, Derevnina L, Sandhu K, Karaoglu H, Elmansour H, Dracatos P, Singh D (2015) Leaf Rust of Cultivated Barley: Pathology and Control. Annu Rev Phytopathol 53:565–589
Parlevliet JE, Kuiper HJ (1977) Partial resistance of barley to leaf rust, Puccinia hordei. IV. Effect of cultivar and development stage on infection frequency. Euphytica 26:249–255
Parlevliet JE, Van Der Beek JG, Pieters R (1981) Presence in Morocco of brown rust, Puccinia hordei, with a wide range of virulence to barley. Cereal Rusts Bull 9:3–8
Peterson RF, Campbell AB, Hannah AE (1948) A diagrammatic scale for estimating rust intensity of leaves and stem of cereals. Can J Res 26:496–500
Qualset C (1975) Sampling germplasm in a center of diversity: an example of disease resistance in Ethiopian barley. Crop genetic resources for today and tomorrow pp 81–96
R Core Team (2018) R: A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria, https://www.R-project.org/
Rehman S, Amouzoune M, Hiddar H, Aberkane H, Benkirane R, Filali-Maltouf A, Al-Jaboobi M, Acqbouch L, Tsivelikas A, Verma R, Kehel Z, Birouk A, Amri A (2020) Traits discovery in Hordeum vulgare sbsp. spontaneum accessions and in lines derived from interspecific crosses with wild Hordeum species for enhancing barley breeding efforts. Crop Sci. https://doi.org/10.1002/csc2.20360
Rothwell CT, Singh D, Dracatos PM, Park RF (2020) Inheritance and characterization of Rph27: a third race-specifc resistance gene in the barley cultivar Quinn. Phytopathology 110:1067–1073
Russell JR, Baum M, Ceccarelli S, Close TM, Grando S, Hayes PM, Matus I, Marshall DF, Del Poza A, von Korff Schmising M, Waugh R (2008) Genomic dissection of tolerance to drought stress in wild barley. 10th International Barley Genetics Symposium, Alexandria, Egypt, 5–10 April 2008
Sandhu KS, Singh D, Park RF (2014) Characterising seedling and adult plant resistance to Puccinia hordei in Hordeum vulgare. Ann Appl Biol 165:117–129
Singh R (1992) Genetic association of leaf rust resistance gene Lr34 with adult plant resistance to stripe rust in bread wheat. Phytopathol 82:835–838
Singh D, Dracatos P, Derevnina L, Zhou MX, Park RF (2015) Rph23: A new designated additive adult plant resistance gene to leaf rust in barley on chromosome 7H. Plant Breed 134:62–69
Stakman EC, Stewart DM, Loegering WQ (1962) Identification of physiologic races of Puccinia graminis var. tritici. USDA Agric Res Serv E617:5–53
Steffenson BJ, Jin Y, Griffey CA (1993) Pathotypes of Puccinia hordei with virulence for the barley leaf rust resistance gene Rph7 in the United States. Plant Dis 77:867–869
Street K, Mackay M, Zuev E, Kaur N, El Bouhssini M, Konopka J, Mitrofanova O (2008) Diving into the genepool: a rational system to access specific traits from large germplasm collections. In: Appels R, Eastwood R, Lagudah E, Langridge P, Mackay M, Mcintyre L, Sharp P(eds) Proceedings of the 11th International Wheat Genetics Symposium , pp 28–31. Brisbane, Australia: Sydney University Press
Stubbs RW, Prescot JM, Saari EE, Dubi HJ (1986) Cereal Disease Methodology Manual. Mexico, Centro Internacional de Mejoramiento de Maizey Trigo (CIMMYT), p 46
Stuthman DD, KJ, Leonard J, Miller-Garvin, (2007) Breeding Crops for Durable Resistance to Disease. Advances in Agronomy, Academic Press 95:319–367
Sun Y (2007) Study of Puccinia hordei and its host resistances in Hordeum vulgare. PhD Thesis, North Dakota State Univ, Fargo, ND
Upadhyaya HD, Oritz R (2001) A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor Appl Genet 102:1292–1298
USDA United States Department of Agriculture Foreign Agricultural Service (FAS), Ministry of Agriculture, Rabat. (2018). (https://gain.fas.usda.gov/RecentGAIN/ Publications/ Grainand/Feed/AnnualRabatMorocco_3–31–2018.pdf)
Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th edn. Springer, New York
Woldeab G, Fininsa C, Singh H, Yuen J (2006) Virulence spectrum of Puccinia hordei in barley production systems in Ethiopia. Plant Pathol 55(3):351–357
Xu Y (2010) Plant genetic resources: Management, evaluation and enhancement. In: Molecular plant breeding. Wallingford, UK, CAB International, 15–194
Yu X, Kong HY, Meiyalaghan V, Casonato S, Chng S, Jones EE, Butler RC, Pickering R, Johnston PA (2018) Genetic mapping of a barley leaf rust resistance gene Rph26 introgressed from Hordeum bulbosum. Theor Appl Genet 131:2567–2580. https://doi.org/10.1007/s00122-018-3173-8
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Ziems LA, Hickey LT, Platz GJ, Franckowiak JD, Dracatos PM, Singh D, Park RF (2017) Characterization of Rph24: a gene conferring adult plant resistance to Puccinia hordei in barley. Phytopath PHYTO-08–16–0295-R
Acknowledgements
The authors would like to thank Dr. Kenneth Street for his help in developing FIGS subset and Mr. Amer El-Omrani, the technician at Sidi Allal Tazi experimental station for his help with field experiments.
Funding
This study is supported financially through GIZ-attributed funding to ICARDA genebank and the CAIGE-GRDC-ICA00010 project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The writers declare that they have no known conflicting financial interests or personal relations that may have had an impact on the work presented in this article.
Human and animal rights
This work does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10722_2021_1268_MOESM1_ESM.xlsx
List of 188 accessions from the reference set constructed within the Generation Challenge Program (GCP) used in the study and relevant information such as IG #, Origin, and collection site (XLSX 18 kb)
10722_2021_1268_MOESM2_ESM.xlsx
List of 86 barley accessions selected using filtering approach of the Focused Identification of Germplasm Strategy (FIGS_LR) and relevant information such as IG #, Origin, and collection site. (XLSX 11 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amouzoune, M., Amri, A., Benkirane, R. et al. Mining and predictive characterization of resistance to leaf rust (Puccinia hordei Otth) using two subsets of barley genetic resources. Genet Resour Crop Evol 69, 839–853 (2022). https://doi.org/10.1007/s10722-021-01268-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01268-4