Abstract
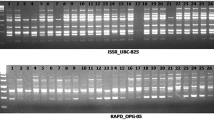
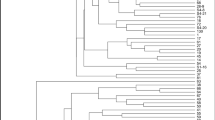
Curcuma caesia Roxb. belonging to Zingiberaceae family is a rhizomatous perennial herb consisting of bluish black rhizome and having vast medicinal and pharmaceutical applications. It is considered as an endangered medicinal plant and native to India. The aim of this study is to assess the molecular diversity among the high rhizome essential oil yielding lines of C. caesia (> 1.5% w/w). The molecular diversity analysis was done by using 45 simple sequence repeat markers amongst the 78 high essential oil yielding lines of C. caesia. The overall percentage of polymorphism amongst the lines was found to be 53%. AMOVA analysis showed 20% and 80% variations among and within the population respectively. Dendrogram constructed using Unweighted Pair Group with Arithmetic Mean (UPGMA) for those high essential oil yielding lines sorted into five major clusters according to their different geographical locations. These five clusters were confirmed using principle component analysis (PCA), based on the Jaccard’s similarity coefficient. The results revealed that these microsatellite markers could be used as successful tool to differentiate the genetic makeup of the crop. Molecular diversity of C. caesia has not been studied till now. So, this is the first scientific report on molecular diversity analysis which may help in the further crop improvement programme and germplasm conservation also.


Similar content being viewed by others
References
Amalraj VA, Velayudhan KC, Muralidharan VK (1989) A note on the anomalous flowering behavior in Curcuma caesia (Zingiberaceae). J Bombay Nat Hist Soc 86:278–279
Baghel SS, Baghel RS, Sharma K, Sikarwar I (2013) Pharmacological activities of Curcuma caesia. Int J Green Pharm 7(1):1–5
Banerjee A, Nigam SS (1976) Antifungal activity of the essential oil of Curcuma caesia Roxb. Indian J Med Res 64:1318–1321
Baruah J, Pandey SK, Begum T, Sarma N, Paw M, Lal M (2019) Molecular diversity assessed amongst high dry rhizome recovery Ginger germplasm (Zingiber officinale Roscoe) from NE-India using RAPD and ISSR markers. Ind Crop Prod 129:463–471
Baruah J, Pandey SK, Sarmah N, Lal M (2019) Assessing molecular diversity among high capsaicin content lines of Capsicum chinense Jacq. using simple sequence repeat marker. Ind Crop Prod 141:111769. https://doi.org/10.1016/j.indcrop.2019.111769
Borah A, Paw M, Gogoi R, Loying R, Sarma N, Munda S, Pandey SK, Lal M (2019) Chemical composition, antioxidant, anti-inflammatory, anti-microbial and in-vitro cytotoxic efficacy of essential oil of Curcuma caesia Roxb. leaves: an endangered medicinal plant of North East India. Ind Crop Prod 129:448–454
Borah A, Kumar D, Paw M, Begum T, Lal M (2020) A review on ethnobotany and promising pharmacological aspects of an endangered medicinal plant, Curcuma caesia Roxb. Turk J Bot 44:205–213. https://doi.org/10.3906/bot-1910-33
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Chopra RN, Chopra IC, Handa KL, Kapur LD (1958) Indigenous drugs of India. U.N. Dhur & Sons Pvt. Ltd., Calcutta
Das S, Mondal P, Zaman MK (2013) Curcuma caesia Roxb and its medicinal uses: a review. IJRPC 6:117–124
Donipati P, Sreeramulu HS (2015) Preliminary phytochemical screening of Curcuma caesia. Int J Curr Microbiol Appl Sci 4:30–34
Dosoky NS, Setzer WN (2018) Chemical composition and biological activities of essential oils of Curcuma species. Nutrients 10(9):1196. https://doi.org/10.3390/nu10091196
Fathima SN, Ahmad SV, Kumar BR (2015) Evaluation of in vitro thrombolytic activity of ethanolic extract of Curcuma caesia rhizomes. Int J Pharm Res Rev 4(11):50–54
Garg SC, Jain RK (1998) Antimicrobial efficacy of essential oil from Curcuma caesia. Ind J Microbiol 38:169–170
Gaur PM, Slinkard AE (1990) Genetic control and linkage relations of additional isozyme markers in Chickpea. Theor Appl Genet 80:648–656
Han Z, Wang C, Song X, Guo W, Gou J, Li C, Chen X, Zhang T (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST-SSR in allotetraploid cotton. Theor Appl Genet 112:430–439
Hasan M, Seyis F, Badani AG, Pons-Kuhnemann J, Friedt W, Lu¨hs W, Snowdon RJ, (2006) Analysis of genetic diversity in the Brassica napus L. gene pool using SSR markers. Genet Resour Crop Evol 53:793–802
Ismail NA, Rafii MY, Mahmud TMM, Hanafi MM, Miah G (2019) Genetic diversity of Torch Ginger (Etlingera elatior) germplasm revealed by ISSR and SSR markers. Bio Med Res Int. https://doi.org/10.1155/2019/5904804
Jain A, Parihar DK (2019) Molecular marker based genetic diversity study of wild, cultivated and endangered species of Curcuma from Chhattisgarh region for in situ conservation. Biocatal Agric Biotechnol. https://doi.org/10.1016/j.bcab.2019.101033
Karmakar I, Saha P, Sarkar N, Bhattacharya S, Haldar PK (2011) Neu-ropharmacological assessment of Curcuma caesia Roxb. rhizome in experimental animal models. Orient Pharm Exp Med 11:251–255
Khare CP (2007) Indian medicinal plants: an illustrated dictionary. Springer, New Delhi
Krishnaraj M, Manibhushanrao K, Mathivanan N (2008) Antimicrobial activity of Curcuma caesia Roxb. Proc Andhara Pradesh Acad Sci 12:280–283
Lian CL, Wadud MA, Geng Q, Shimatani K (2006) An improved technique for isolating co dominant compound microsatellite. Markers J Plant Res 19:415–441
Liu Y, Roy SS, Nebie RHC, ZhangY NMG (2013) Functional food quality of Curcuma caesia, Curcuma zedoaria and Curcuma aeruginosa endemic to North-eastern India. Plant Foods Hum Nutr 68:72–77
Mahanta BP, Sut D, Kemprai P, Paw M, Lal M, Haldar S (2020) A 1 H-NMR spectroscopic method for the analysis of thermolabile chemical markers from the essential oil of black turmeric (Curcuma caesia) rhizome: application in post-harvest analysis. Phytochem Anal 31(1):28–36. https://doi.org/10.1002/pca.2863
Mamatha K (2016) Studies on genetic diversity in turmeric (Curcuma longa L.) genotypes using morphological and molecular markers. Dissertation, College of Horticulture, Rajendranagar, Hyderabad, India
Mann A, Dahiya BS, Boora KS, Nandwal AS, Vashist U (2010) PCR-Based genetic diversity in Chickpea (Cicer arietinum L.) genotypes using random primers. IUP J Genet Evol 3:1–12
Mitchess SE, Kresovich S, Jester CA, Hernandez CJ, Szewc- Mcfadden AK (1997) Application of multiplex PCR and fluorescence-based semi-automated allele sizing technology genotyping plant genetic resources. Crop Sci 37:617–624
Mohanty S, Panda MK, Acharya L, Nayak S (2014) Genetic diversity and gene differentiation among ten species of Zingiberaceae from Eastern India. 3 Biotech 4(4):383–390. https://doi.org/10.1007/s13205-013-0166-9
Mukunthan KS, Kumar NVA, Balaji S, Trupti NP (2014) Analysis of essential oil constituents in rhizome of Curcuma caesia Roxb. from South India. J Essent Oil Bear Plant 17:647–651
Nadeem MA, Nawaz MA, Shahid MQ, Dogan Y, Comertpay G, Yildiz M, Baloch FS (2017) DNA markers in plant breeding current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32:261–285
Pandey AK, Chowdhury AR (2003) Volatile constituents of the rhizome oil of Curcuma caesia Roxb. from central India. Flavour Fragr J 18:463–465
Pandey D, Gupta AK (2014) Antibacterial efficacy of Curcuma caesia from Bastar district of Chattisgarh, India. Int J Pharm Sci Res 5:2294–2301
Paliwal A (2016) Effectiveness of STMS markers for generating unique DNA profiles in Chickpea (Cicer arietinum L.) germplasm. Int J New Technol Res 2:79–83
Paliwal P, Pancholi SS, Patel RK (2011) Pharmacognostic parameters for evaluation of the rhizomes of Curcuma caesia. J Adv Pharm Technol Res 2:56–61
Paw M, Gogoi R, Sarma N, Pandey SK, Borah A, Begum T, Lal M (2020) Study of anti-oxidant, anti-inflammatory, genotoxicity, antimicrobial activities and analysis of different constituents found in rhizome essential oil of Curcuma caesia Roxb., collected from North East India. Curr Pharm Biotechnol 21:403–413. https://doi.org/10.2174/1389201020666191118121609
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Raut JS, Karuppayil SM (2014) A status review on the medicinal properties of essential oils. Ind Crops Prod 62:250–264. https://doi.org/10.1016/j.indcrop.2014.05.055
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system Version 2.1. Exeter Software, Setauket
Sahoo A, Jena S, Kar B, Sahoo S, Ray A, Singh S, Nayak S (2017) EST-SSR marker revealed effective over biochemical and morphological scepticism towards identification of specific turmeric (Curcuma longa L.) cultivars. 3 Biotech. https://doi.org/10.1007/s13205-017-0701-1
Sahu B, Kenwat R, Chandrakar S (2016) Medicinal value of Curcuma cassiaRoxb: an overview. UKJPB 4:69–74
Sahu R, Saxena J (2018) Bioactive compound from rhizome part of Curcuma caesia. Int J Pharm Sci 3:6–8
Sarangthem K, Haokip MJ (2010) Bioactive components in Curcuma caesia Roxb. grown in Manipur. Bioscan 5:113–115
Sasikumar B (2005) Genetic resource of Curcuma: diversity, characterization and utilization. Plant Genet Res 3:230–251
Senan S, Kizhakayil D, Sheeja TE, Sasikumar B, Bhat AI, Parthasarathy VA (2013) Novel polymorphic microsatellite markers from turmeric, Curcuma longa L. (Zingiberaceae). Acta Bot Croat 72(2):407–412. https://doi.org/10.2478/botcro-2013-0002
Sharma N, Verma PP, Murthy SN (2017) Pharmacognostical evaluation and conservation of threatened species Curcuma caesia Roxb. Int J Ayurvedic Med 8:68–72
Sigrist MS, Pinheiro JB, Azevedo Filho JA, Zucchi MI (2011) Genetic diversity of turmeric germplasm (Curcuma longa; Zingiberaceae) identified by microsatellite markers. Genet Mol Res 10:419–428
Sigrist MS, Pinheiro JB, Azevedo-Filho JA, Colombo CA, Bajay MM, Lima PF, Zucchi MI (2009) Development and characterization of microsatellite markers for turmeric (Curcuma longa). Plant Breed 129:570–573
Siju S, Dhanya K, Syamkumar S, Sheeja TE, Sasikumar B, Bhat AI, Parthasarathy VA (2010) Development, characterization and utilization of genomic microsatellite markers in turmeric (Curcuma longa L.). Biochem Syst Ecol 38:641–646. https://doi.org/10.1016/j.bse.2010.08.006
Singh BD (2001) Fundamentals of genetics. Kalyani Publishers, New Delhi
Singh S, Panda MK, Nayak S (2012) Evaluation of genetic diversity in turmeric (Curcuma longa L.) using RAPD and ISSR markers. Ind Crop Prod 37:284–291. https://doi.org/10.1016/j.indcrop.2011.12.022
Singh TJ, Patel RK, Patel SN, Patel PA (2018) Molecular diversity analysis in Turmeric (Curcuma longa L.) using SSR markers. Int J Curr Microbiol Appl Sci 7:552–560
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Tag H, Das AK, Loyi H (2007) Anti inflammatory plant used by Khamti tribes of Lohit district in Arunachal Pradesh. Nat Prod Radiance 4:334–340
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
The Plant List (2013) Version 1.1. Published on the Internet. http://www.theplantlist.org/. Accessed 1 Sept (2018)
Valdes AM, Slatkin M, Freimer NB (1993) Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics 133:737–749
Vieira MLC, Santini L, Diniz AL, Munhoz CF (2016) Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol 39(3):312–328
Xia X, Luan LL, Qin G et al (2017) Genome-wide analysis of SSR and ILP markers in trees: diversity profiling, alternate distribution, and applications in duplication. Sci Rep 7(1):17902. https://doi.org/10.1038/s41598-017-17203-6
Yeh FC, Yang RC, Boyle T (2000) POPGENE 1.32: a free program for the analysis of genetic variation among and within populations using co-dominant and dominant markers. Department of Ren. Res., Uni Alberta, Canada
Zheng WH, Zhuo Y, Liang L, Ding WY, Liang LY, Wang XF (2015) Conservation and population genetic diversity of Curcuma wenyujin (Zingiberaceae), a multifunctional medicinal herb. Genet Mol Res 14:10422–10432
Acknowledgements
The authors are grateful to the Director, CSIR-NESIT, Jorhat (Assam) for providing the lab and field facilities and special thanks to The NMPB, Ministry of AYUSH, Govt. of India, and New Delhi for funding this research work in the form of R&D Project Number AS/01/2016-17
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest on the content of manuscript and study undertaken.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Paw, M., Borah, A., Pandey, S.K. et al. Simple sequence repeat marker based genetic diversity assessment amongst high essential oil yielding lines of Curcuma caesia Roxb.. Genet Resour Crop Evol 68, 1345–1358 (2021). https://doi.org/10.1007/s10722-020-01066-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-01066-4