Abstract
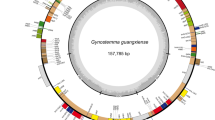
Gardenia jasminoides, a well-known traditional Chinese herb, has been extensively utilized for its being as stable pigment in food industry and as rich source of crocin in treatment of various cancers and cardiovascular disease. Nevertheless, molecular data shortage of the organelle and nuclear genome vastly limits further biological research on G. jasminoides. In this study, complete chloroplast (cp) genome and large-scale nuclear genome sequences of G. jasminoides were initially generated by genome survey sequencing, and comprehensive genomic structure and comparison analyses were performed. The results showed that G. jasminoides cp genome displayed a typical quadripartite organization and was sized in 154,921 bp with 37.5% GC content. It contained 132 genes, 18 out of which were duplicated in the inverted repeat regions (IRs). Fifty long repeats (motif > 10 bp) and 27 simple sequence repeats were detected, and 30 usage-biased codons and 58 RNA-editing sites were predicted in G. jasminoides cp genome. The G. jasminoides cp genome was similar to those related species in Rubiaceae in terms of gene order and IR borders and a sister relationship of Gardenia and Coffea is strongly supported. Furthermore, the nuclear genome size of G. jasminoides was estimated to be 562.9 Mb, with heterozygosity rate being of 0.68%, repeat proportion of 51.6% and GC content of 36.44%. Overall, the data resources and findings generated in the present study will shed new light on the molecular and breeding studies on this important crocin-producing plant in the future.







Similar content being viewed by others
References
Amiryousefi A, Hyvönen J, Poczai P (2018) IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34:3030–3031
Beaulieu JM, Smith SA, Leitch IJ (2010) On the tempo of genome size evolution in angiosperms. J Botany 2010:989152
Bhandari PR (2015) Crocus sativus L. (saffron) for cancer chemoprevention: a mini review. J Tradit Complement Med 5:81–87
Bock R (2014) Genetic engineering of the chloroplast: novel tools and new applications. Curr Opin Biotechnol 26:7–13
Bock DG, Kane NC, Ebert DP, Rieseberg LH (2014) Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: neither from Jerusalem nor an artichoke. New Phytol 201:1021–1030
Bremer B, Eriksson T (2009) Time tree of Rubiaceae: phylogeny and dating the family, subfamilies, and tribes. Int J Plant Sci 170:766–793
Chen T, Luo XR, Zhu H, Charlotte MT, Friedrich E, Henrik L, Michele F, Christian P, Wu ZY, Raven PH (2011) Rubiaceae. Science Press, Beijing, p 570
Chen X, Zhou J, Cui Y, Wang Y, Duan B, Yao H (2018) Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front Pharmacol 9:695
Corneille S, Lutz K, Maliga P (2000) Conservation of RNA editing between rice and maize plastids: are most editing events dispensable? Mol Gen Genet 264:419–424
Daniell H, Lin C-S, Yu M, Chang W-J (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17:134
de Santana Lopes A, Pacheco TG, dos Santos KG, do Nascimento Vieira L, Guerra MP, Nodari RO, de Souza EM, de Oliveira Pedrosa F, Rogalski M (2018) The Linum usitatissimum L. plastome reveals atypical structural evolution, new editing sites, and the phylogenetic position of Linaceae within Malpighiales. Plant Cell Rep 37:307–328
Deng SY, Wang XR, Zhu PL, Wen Q, Yang CX (2015) Development of polymorphic microsatellite markers in the medicinal plant Gardenia jasminoides (Rubiaceae). Biochem Syst Ecol 58:149–155
Downie SR, Jansen RK (2015) A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst Botany 40:336–351
Duan R, Huang M, Yang L, Liu Z (2017) Characterization of the complete chloroplast genome of Emmenopterys henryi (Gentianales: Rubiaceae), an endangered relict tree species endemic to China. Conserv Genet Resour 9:459–461
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279
Freyer R, Hoch B, Neckermann K, Maier RM, Kössel H (1993) RNA editing in maize chloroplasts is a processing step independent of splicing and cleavage to monocistronic mRNAs. Plant J 4:621–629
Frusciante S, Diretto G, Bruno M, Ferrante P, Giuliano G (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci 5:12246–12251
Gaber MK, Barakat AA (2019) Micropropagation and somatic embryogenesis induction of Gardenia jasminoides plants. Alex Sci Exch J 40:190–202
Han J, Zhang W, Cao H, Chen S, Wang Y (2007a) Genetic diversity and biogeography of the traditional Chinese medicine, Gardenia jasminoides, based on AFLP markers. Biochem Syst Ecol 35:138–145
Han J, Chen S, Zhang WS, Wang Y (2007b) Molecular ecology of Gardenia jasminoides authenticity. Chin J Appl Ecol 17:2385–2388
Hirose T, Sugiura M (1997) Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J 16:6804–6811
Hosseinzadeh H, Sadeghnia HR, Ghaeni FA, Motamedshariaty VS, Mohajeri SA (2012) Effects of saffron (Crocus sativus L.) and its active constituent, crocin, on recognition and spatial memory after chronic cerebral hypoperfusion in rats. Phytother Res 26:381–386
Huang Y, Yang Z, Huang S, An W, Li J, Zheng X (2019) Comprehensive analysis of Rhodomyrtus tomentosa chloroplast genome. Plants 8:89
Ji A, Jing J, Zhichao X, Ying L, Wu B, Fengming R, Chunnian H, Jie L, Kaizhi H, Jingyuan S (2017) Transcriptome-guided mining of genes involved in crocin biosynthesis. Front Plant Sci 8:518
Jiao W-B, Schneeberger K (2017) The impact of third generation genomic technologies on plant genome assembly. Curr Opin Plant Biol 36:64–70
Kahlau S, Aspinall S, Gray JC, Bock R (2006) Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J Mol Evol 63:194–207
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kersey PJ (2019) Plant genome sequences: past, present, future. Curr Opin Plant Biol 48:1–8
Kowallik KV (2018) Origin and evolution of chloroplasts: current status and future perspectives. In: Schenk HEA, Herrmann RG, Jeon KW, Müller NE, Schwemmler W (eds) Eukaryotism and symbiosis. Springer, Berlin, pp 3–23
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642
Lai CL, Yang JS (1999) Production of crocin by fruit callus culture of Gardenia jasminoides Ellis. Food Biotechnol 13:209–216
Lei L, Wang Y, Zhao A-M, Zhu P-L, Zhou SL (2009) Genetic relationship of Gardenia jasminoides among plantations revealed by ISSR. Chin Tradit Herb Drugs 40:117–120
Li C, Lin F, An D, Wang W, Huang R (2018) Genome sequencing and assembly by long reads in plants. Genes 9:6
Li H-T, Yi T-S, Gao L-M, Ma P-F, Zhang T, Yang J-B, Gitzendanner MA, Fritsch PW, Cai J, Luo Y (2019) Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants 5:461
Lu M, An H, Li L (2016) Genome survey sequencing for the characterization of the genetic background of Rosa roxburghii Tratt and leaf ascorbate metabolism genes. PLoS ONE 11:e0147530
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18
Ma P-F, Zhang Y-X, Zeng C-X, Guo Z-H, Li D-Z (2014) Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst Biol 63:933–950
Mai N, Terasaka K, Owaki M, Sota M, Inukai T, Nagatsu A, Mizukami H (2012) UGT75L6 and UGT94E5 mediate sequential glucosylation of crocetin to crocin in Gardenia jasminoides. FEBS Lett 586:1055–1061
Maier RM, Zeitz P, Kössel H, Bonnard G, Gualberto JM, Grienenberger JM (1996) RNA editing in plant mitochondria and chloroplasts. In: Filipowicz W, Hohn T (eds) Post-transcriptional control of gene expression in plants. Springer, Berlin, pp 343–365
Marc L, Oliver D, Sabine K, Ralph B (2013) OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:W575
Mei Z, Zhou B, Wei C, Cheng J, Imani S, Chen H, Fu J (2015) Genetic authentication of Gardenia jasminoides Ellis var. grandiflora Nakai by improved RAPD-derived DNA markers. Molecules 20:20219–20229
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), New Orleans, LA, USA, 14 November, pp 1–8
Mower JP (2009) The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res 37:W253–W259
Peng K, Yang L, Zhao S, Chen L, Zhao F, Qiu F (2013) Chemical constituents from the fruit of Gardenia jasminoides and their inhibitory effects on nitric oxide production. Bioorg Med Chem Lett 23:1127–1131
Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom 8:174
Razavi BM, Hosseinzadeh H, Movassaghi AR, Imenshahidi M, Abnous K (2013) Protective effect of crocin on diazinon induced cardiotoxicity in rats in subchronic exposure. Chemico-Biol Interact 203:547–555
Robinson AC, Castañeda CA, Schlessman JL (2014) Structural and thermodynamic consequences of burial of an artificial ion pair in the hydrophobic interior of a protein. Proc Natl Acad Sci 111:11685–11690
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Saarela JM, Burke SV, Wysocki WP, Barrett MD, Clark LG, Craine JM, Peterson PM, Soreng RJ, Vorontsova MS, Duvall MR (2018) A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions. PeerJ 6:e4299
Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H (2007) The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol J 5:339–353
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689
Sheu S, Hsin W (1998) HPLC separation of the major constituents of gardeniae fructus. J Separ Sci 21:523–526
Sloan DB, Taylor DR (2010) Testing for selection on synonymous sites in plant mitochondrial DNA: the role of codon bias and RNA editing. J Mol Evol 70:479–491
Sonah H, Deshmukh RK, Singh VP, Gupta DK, Singh NK, Sharma TR (2011) Genomic resources in horticultural crops: status, utility and challenges. Biotechnol Adv 29:199–209
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017) GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45:W6–W11
Tsanakas GF, Manioudaki ME, Economo AS, De Kalaitzis P (2014) novo transcriptome analysis of petal senescence in Gardenia jasminoides Ellis. BMC Genom 15:554
Tsudzuki T, Wakasugi T, Sugiura M (2001) Comparative analysis of RNA editing sites in higher plant chloroplasts. J Mol Evol 53:327–332
Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, Gurtowski J, Schatz MC (2017) GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33:2202–2204
Wang WC, Chen SY, Zhang XZ (2016) Chloroplast genome evolution in Actinidiaceae: clpP loss, heterogenous divergence and phylogenomic practice. PLoS ONE 11:e0162324
Wang W, Chen S, Zhang X (2018a) Whole-genome comparison reveals heterogeneous divergence and mutation hotspots in chloroplast genome of Eucommia ulmoides Oliver. Int J Mol Sci 19:1037
Wang W, Chen S, Zhang X (2018b) Whole-genome comparison reveals divergent IR borders and mutation hotspots in chloroplast genomes of herbaceous bamboos (Bambusoideae: Olyreae). Molecules 23:1537
Wang L, Xing H, Yuan Y, Wang X, Saeed M, Tao J, Feng W, Zhang G, Song X, Sun X (2018c) Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS ONE 13:e0194372
Wang S, Chen S, Liu C, Liu Y, Zhao X, Yang C, Qu G-Z (2019a) Genome survey sequencing of Betula platyphylla. Forests 10:826
Wang R, Fan J, Chang P, Zhu L, Zhao M, Li L (2019b) Genome survey sequencing of Acer truncatum Bunge to identify genomic information, simple sequence repeat (SSR) markers and complete chloroplast genome. Forests 10:87
Wicke S, Schneeweiss GM, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76:273–297
Wu Y, Wu R, Zhang B, Jiang T, Li N, Qian K, Zhang J (2012) Epigenetic instability in genetically stable micropropagated plants of Gardenia jasminoides Ellis. Plant Growth Regul 66:137–143
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Xu Z-C, Pu X-D, Gao R-R, Demurtas OC, Fleck SJ, Richter M et al (2020) Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol 18:63
Yan C, Yang ZL, Zhang LH, Liu SJ, Zhang XT (2011) Determination of geniposide, crocin and crocetin in different processing products of fructus gardeniae by HPLC-ELSD. J Chin Med Mater 34:687–690
Yang Z, Huang Y, An W, Zheng X, Huang S, Liang L (2019) Sequencing and structural analysis of the complete chloroplast genome of the medicinal plant Lycium chinense Mill. Plants 8:87
Zhang Y, Zhang J-W, Yang Y, Li X-N (2019) Structural and comparative analysis of the complete chloroplast genome of a mangrove plant: Scyphiphora hydrophyllacea Gaertn. f. and related Rubiaceae species. Forests 10:1000
Zhao H, Yang L, Peng Z, Sun H, Yue X, Lou Y, Dong L, Wang L, Gao Z (2015) Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci Rep 5:8018
Zhu T, Wang L, You FM, Rodriguez JC, Deal KR, Chen L et al (2019) Sequencing a Juglans regia × J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic Res 6:1–16
Acknowledgements
We would like to thank Qian Liu from BioMarker for the help in DNA sequencing experiment.
Funding
This work is supported by National Natural Science Foundation of China (Grant No. 31600173), Xinglin Talent Funding of Guangzhou University of Chinese Medicine (A1-AFD018181Z3949), Innovation Project for Forestry Science and Technology in Guangdong (2019KJCX012, 2020KYXM03, 2020KJCX010), and Excellent Doctor Fund of Zhongkai University of Agriculture and Engineering (KA180581235).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: W-CW, X-ZZ. Performed the experiments: W-CW, F-QS, XD, Y-WL. Analyzed the data: W-CW, S-YC, X-ZZ. Contributed reagents/materials/analysis tools: Y-WL, S-YC, Y-QL, WG, Q-BJ, HL, X-ZZ. Wrote the paper: W-CW, X-ZZ. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, W., Shao, F., Deng, X. et al. Genome surveying reveals the complete chloroplast genome and nuclear genomic features of the crocin-producing plant Gardenia jasminoides Ellis. Genet Resour Crop Evol 68, 1165–1180 (2021). https://doi.org/10.1007/s10722-020-01056-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-01056-6