Abstract
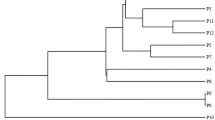
Seedling germplasm resources are important for mango breeding. Genetic diversity analyses based on molecular data can provide useful information for germplasm management and varietal characterization. Start codon targeted (SCoT) markers are efficient and highly polymorphic markers for phylogenetic relationship analyses of multiple species and were used in the present study to determine genetic relatedness and variability among 168 mango (Mangifera indica L.) germplasm resources. Forty-five SCoT primers generated unambiguous and reproducible bands and a total of 337 fragments, with a mean of 7.49 fragments per primer. Of these, 244 (72.4%) were polymorphic. The number of polymorphic bands varied from 1 (SCoT55) to 10 (SCoT4 and 52), with an average of 5.42 amplicons per primer. The average polymorphism information content, Nei’s genetic diversity, Shannon’s information index and marker index were 0.823, 0.426, 0.603 and 4.460, respectively. A dendrogram generated using the unweighted pair-group method with arithmetic mean delineated the 168 germplasm resources into two major clusters. Analysis of molecular variance revealed higher genetic diversity (89%) within each population than among the populations (11%). Most mango seedling germplasm resources grouped with their potential parents, and genetic differences were also detected. Thirty-four seedling germplasm resources were identified as potential parents or sister lines. These results demonstrate that SCoT markers are useful for cultivar identification and genetic diversity analyses of mango cultivars. This genetic information will support germplasm management and cultivar improvement.





Similar content being viewed by others
References
Ahmad R, Anjum MA, Malik W (2018) Characterization and evaluation of mango germplasm through morphological, biochemical, and molecular markers focusing on fruit production: an overview. Mol Biotechnol. https://doi.org/10.1007/s12033-018-0129-9
Amom T, Nongdam P (2017) The use of molecular marker methods in plants: a review. Int J Curr Res Rev 9(17):1–7
Ariffin Z, Shafie M, Sah MSM, Idris S, Hashim N (2015) Genetic diversity of selected Mangifera species revealed by inter simple sequence repeats markers. Int J Biodivers. https://doi.org/10.1155/2015/458237
Azmat MA, Khan AA, Khan IA, Rajwana IA, Cheema HMN, Khan AS (2016) Morphological characterization and SSR based DNA fingerprinting of elite commercial mango cultivars. Pak J Agr Sci 53(2):321–330
Bajpai A, Muthukumar M, Ahmad I, Ravishankar KV, Parthasarthy VA, Sthapit BR, Rao RV, Sharma JP, Rajan S (2016) Molecular and morphological diversity of locally grown heirloom mango varieties of North India. J Environ Biol 37(2):221–228
Bompard JM, Schnell RJ (1997) Taxonomy and systematics. In: Litz RE (ed) The mango: botany, production and uses. CAB International, Wallingford, pp 21–47
Chen JZ (2011) Fruit tree cultivation (southern version). China Agricultural Press, Beijing, pp 198–219
Chen FY, Liu JH (2014) Germplasm genetic diversity of Myrica rubra, in Zhejiang province studied using inter-primer binding site and start codon-targeted polymorphism markers. Sci Hortic 170(3):169–175
Chhajer S, Jukanti AK, Kalia RK (2017) Start codon targeted (SCoT) polymorphism-based genetic relationships and diversity among populations of Tecomella undulata (Sm.) Seem—an endangered timber tree of hot arid regions. Tree Genet Genomes 13(4):84
Collard BCY, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Rep 27(1):86–93
Deng LB, Liang QZ, He XH, Luo C, Chen H, Qin ZS (2015) Investigation and analysis of genetic diversity of diospyros germplasms using scot molecular markers in Guangxi. PLoS ONE 10(8):e0136510
Dillon NL, Bally ISE, Wright CL, Hucks L, Innes DJ, Dietzgen RG (2013) Genetic diversity of the Australian national mango genebank. Sci Hortic 150:213–226
Dong YC, Liu X (2006) Crops and their wild relatives in China [vol. fruit crops]. China Agricultural Press, Beijing, pp 583–603
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics 131:479–491
Feng SG, He RF, Yang S, Chen Z, Jiang MY, Lu JJ, Wang HZ (2015) Start codon targeted (SCoT) and target region amplification polymorphism (TRAP) for evaluating the genetic relationship of dendrobium species. Gene 567(2):182–188
Gajera HP, Bambharolia RP, Domadiya RK, Patel SV, Golakiya BA (2014) Molecular characterization and genetic variability studies associated with fruit quality of indigenous mango (Mangifera indica L.) cultivars. Plant Syst Evol 300(5):1011–1020
Gitahi R, Kasili R, Kyallo M, Kehlenbeck K (2016) Diversity of threatened local mango landraces on smallholder farms in Eastern Kenya. For Trees Livelihoods 25(4):239–254
Gorji AM, Poczai P, Polgar Z, Taller J (2011) Efficiency of arbitrarily amplified dominant markers (SCOT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am J Potato Res 88(3):226–237
Guo DL, Zhang JY, Liu CH (2012) Genetic diversity in some grape varieties revealed by SCoT analyses. Mol Biol Rep 39(5):5307–5313
Hao J, Jiao KL, Yu CL, Guo H, Zhu YJ, Yang X, Zhang SY, Zhang L, Feng SG, Song YB, Dong M, Wang HZ, Shen CJ (2018) Development of SCoT-based SCAR marker for rapid authentication of Taxus Media. Biochem Genet 56:255–266
He XH, Li YR, Guo YZ, Tang ZP, Li RB (2005) Genetic analysis of 23 mango cultivar collection in Guangxi province revealed by ISSR. Mol Plant Breed 3(6):829–834
He XH, Guo YZ, Li YR, Ou SJ (2007a) Assessment of the genetic relationship and diversity of mango and its relatives by cplSSR marker. Agric Sci China 6(2):137–142
He XH, Li YR, Guo YZ, Ou SJ, Li RB (2007b) Identification of closely related mango cultivars by ISSR. Guihaia 27(1):44–47
He SY, Niu JX, Ma JJ (2016) Optimization of start codon targeted polymorphism PCR system and identification of bud mutant in ‘Kuerle Xiangli’. J Fruit Sci 33(11):1337–1346
Jalilian H, Zarei A, Erfani-Moghadam J (2018) Phylogeny relationship among commercial and wild pear species based on morphological characteristics and SCoT molecular markers. Sci Hortic 235:323–333
Karihaloo JL, Dwivedi YK, Archak S, Gaikwad A (2003) Analysis of genetic diversity of Indian mango cultivars using RAPD markers. J Hortic Sci Biotechnol 78(3):285–289
Kashkush K, Fang JG, Tomer E, Hillel J, Lavi U (2001) Cultivar identification and genetic map of mango (Mangifera indica). Euphytica 122(1):129–136
Khan AS, Ali S, Khan IA (2015) Morphological and molecular characterization and evaluation of mango germplasm: an overview. Sci Hortic 194:353–366
Krishna H, Singh SK (2007) Biotechnological advances in mango (Mangifera indica L.) and their future implication in crop improvement—a review. Biotechnol Adv 25(3):223–243
Kuhn DN, Bally ISE, Dillon NL, Innes D, Groh AM, Rahaman J, Ophir R, Cohen Y, Sherman A (2017) Genetic map of mango: a tool for mango breeding. Front Plant Sci 8:577
Kumar M, Sharma R, Kumar V, Sirohi U, Chaudhary V, Sharma S, Saripalli G, Naresh RK, Yadav HK, Sharma S (2019) Genetic diversity and population structure analysis of Indian garlic (Allium sativum L.) collection using SSR markers. Physiol Mol Biol Plants 25(2):377–386
Lal S, Singh AK, Singh SK, Srivastav M, Singh BP, Sharma N, Singh NK (2017) Association analysis for pomological traits in mango (Mangifera indica L.) by genic-SSR markers. Trees Struct Funct 31(5):1391–1409
Lei XT, Wang JB, Xu XR, Lin SQ (2006) Studies on genetic polymorphism of main mango cultivars via AFLP markers. Acta Hortic Sin 33(4):725–730
Luo C, He XH, Chen H, Ou SJ, Gao MP (2010) Analysis of diversity and relationships among mango cultivars using start codon targeted (SCoT) markers. Biochem Syst Ecol 38(6):1176–1184
Luo C, He XH, Chen H, Ou SJ, Gao MP, Brown JS, Tondo CL, Schnell RJ (2011) Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem Syst Ecol 39(4–6):676–684
Luo C, He XH, Chen H, Hu Y, Ou SJ (2012) Genetic relationship and diversity of Mangifera indica L.: revealed through SCoT analysis. Genet Resour Crop Evol 59(7):1505–1515
Mahjbi A, Baraket G, Oueslati A, Salhi-Hannachi A (2015) Start codon targeted (SCoT) markers provide new insights into the genetic diversity analysis and characterization of Tunisian citrus, species. Biochem Syst Ecol 61:390–398
Mulpuri S, Muddanuru T, Francis G (2013) Start codon targeted (SCoT) polymorphism in toxic and non-toxic accessions of Jatropha curcas L. and development of a codominant SCAR marker. Plant Sci 207:117–127
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Chung G, Baloch FS (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32:261–285
Nazish T, Shabbir G, Ali A, Sami-ul-Allah S, Naeem M, Javed M, Batool S, Arshad H, Hussain SB, Aslam K, Seher R, Tahir M, Baber M (2017) Molecular diversity of Pakistani mango (Mangifera indica L.) varieties based on microsatellite markers. Genet Mol Res 16(2):1–9
Olano CT, Schnell RJ, Quintanilla WE, Campbell RJ (2005) Pedigree analysis of Florida mango cultivars. Proc Fla State Hort Soc 118:192–197
Peakall R, Smouse PE (2006) GenALEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2(3):225–238
Pritchard JK, Stephens MJ, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Rahimi M, Nazari L, Kordrostami M, Safari P (2018) SCoT marker diversity among Iranian Plantago ecotypes and their possible association with agronomic traits. Sci Hortic 233:302–309
Rajesh MK, Sabana AA, Rachana KE, Rahman S, Jerard BA, Karun A (2015) Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through scot analysis. Biotech 5(6):999–1006
Ramessur AD, Ranghoosanmukhiya VM (2011) RAPD marker-assisted identification of genetic diversity among mango (Mangifera indica) varieties in Mauritius. Int J Agric Biol 13(2):167–173
Ravishankar KV, Bommisetty P, Bajpai A, Srivastava N, Dinesh MR (2015) Genetic diversity and population structure analysis of mango (Mangifera indica) cultivars assessed by microsatellite markers. Trees Struct Funct 29(3):775–783
Raza SA, Khan AS, Khan IA, Rajwana IA, Ali S, Khan AA, Rehman A (2017) Morphological and physico-chemical diversity in some indigenous mango (Mangifera indica L.) germplasm of Pakistan. Pak J Agr Sci 54(2):287–297
Rohlf FJ (2000) NTSYS-PC. Numerical taxonomy and multivariate analysis system version 2.10. Exeter Software, Setauket
Samal KC, Jena RC, Swain SS, Das BK, Chand PK (2012) Evaluation of genetic diversity among commercial cultivars, hybrids and local mango (Mangifera indica L.) genotypes of India using cumulative RAPD and ISSR markers. Euphytica 185(2):195–213
Schnell RJ, Ronning CM, Knight RJ (1995) Identification of cultivars and validation of genetic relationships in Mangifera indica L. using RAPD markers. Theor Appl Genet 90(2):269–274
Schnell R, Brown J, Olano C, Meerow AW, Campbell RJ, Kuhn DN (2006) Mango genetic diversity analysis and pedigree inferences for Florida cultivars using microsatellite markers. J Am Soc Hortic Sci 131(2):214–224
Shekhawat JK, Rai MK, Shekhawat NS, Kataria V (2018) Start codon targeted (SCoT) polymorphism for evaluation of genetic diversity of wild population of Maytenus emarginata. Ind Crops Prod 122:202–208
Shi SY, Wu HX, Wang SB, Yao QS, Liu LQ, Wang YC, Ma WH, Zhan RL (2011) Diversity analysis of fruit quality traits in loquat fruit germplasm. Acta Hortic Sin 38(05):840–848
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38:1409–1438
Souza IG, Valente SE, Britto FB, Souza VA, Lima PS (2011) RAPD analysis of the genetic, diversity of mango (Mangifera indica) germplasm in Brazil. Genet Mol Res 10(4):3080–3089
Surapaneni M, Vemireddy LR, Begum H, Reddy BP, Neetasri C, Nagaraju J, Anwar SY, Siddiq EA (2013) Population structure and genetic analysis of different utility types of mango (Mangifera indica L.) germplasm of Andhra Pradesh state of India using microsatellite markers. Plant Syst Evol 299(7):1215–1229
Thumar ST, Gajera HP, Dhaduk HL, Sakure AA (2016) Physiological, qualitative and molecular markers based evaluation of mango cultivars. Indian J Agric Biochem 29(1):80–86
Tsai CC, Chen Y, Chen CH, Weng I, Tsai CM, Lee SR, Lin YS, Chiang YC (2013) Cultivar identification and genetic relationship of mango (Mangifera indica) in Taiwan using 37 SSR markers. Sci Hortic 164:196–201
Wang LX, Wu ZX, Wang JB, Li SP, Jiang CD (2003) Fuzzy cluster analysis of Hainan main mango cultivars according to leaves characters. Chin J Trop Crop 24(4):28–34
Wang JB, Wang LX, Du ZJ, Lei XT, Chen YY, Xu BY (2007) Analysis on the genetic relationship of some mango (Mangifera indica L.) germplasms by ISSR markers. Acta Hortic Sin 34(1):87–92
Wang JY, Wang JB, Wu YT, Huang GD, Zhou JA, Li RW (2009) Analysis of genetic diversity of major accessions of mango (Mangifera indica L.) in Guangxi using SSR markers. Chin J Trop Crop 30(9):1301–1307
Wang M, Ying DS, Wang QF, Li LP, Zhang RL (2015) Genetic diversity analysis and fingerprint construction for mango cultivars in China. J South Agric 46(7):1154–1159
Wu XL, Zheng FQ, Xu CX, Tang WW (2016) Genetic diversity analysis of Shatangju mandarin (Citrus reticulata) by SCoT-PCR. Agric Sci Technol 17(1):34–37+68
Xie JH, Liu CM, Ma WH, Lei XT (2005) Analysis of genetic relationships among mango germplasm by RAPD markers. J Fruit Sci 22(6):649–653
Xiong FQ, Tang RH, Chen ZL, Pan LH, Zhuang WJ (2009) SCoT: a novel gene targeted marker technique based on the translation start codon. Mol Plant Breed 7(3):635–638
Yamanaka N, Hasran M, Xu DH, Tsunematsu H, Idris S, Ban T (2006) Genetic relationship and diversity of four Mangifera species revealed through AFLP analysis. Genet Resour Crop Evol 53(5):949–954
Zhan X, Li P, Hui WK, Deng YW, Gan SM, Sun Y, Zhao XH, Chen XY, Deng XM (2019) Genetic diversity and population structure of Toona ciliata revealed by simple sequence repeat markers. Biotechnol Biotechnol Equip 5:1–9
Zhang YY, Chen SY (1992) Classification of tropical and subtropical fruits. Shanghai Science and Technology Press, Shanghai, pp 85–98
Zhang CX, Ni ZG, Chen HR, Chen YF, Xie DH, Long YQ, Zhang FM (2014) Genetic diversity of mango (Mangifera indica L.) in Nujiang dry-hot valley revealed by morphological characters and AFLP marker. J Plant Genet Resour 15(4):753–758
Acknowledgements
This research was supported by National Natural Science Foundation of China (31660561 and 31860541), Key Research and Development Project in Guangxi (GXKJ-AB17292010), Major Science and Technology Projects in Guangxi (GXKJ-AA17204026-2, GXKJ-AA17204097-3, and GXKJ-AA17204097-10), State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-a201906; SKLCUSA-c201901), Natural Science Foundation of Guangxi Province (2015GXNSFAA139052), Innovation Team of Guangxi Mango Industry Project (nycytxgxcxtd-06-02), Innovation Project of Guangxi Graduate Education (YCSW2018040), and Agricultural Science and Technology Project of Guangxi (201608).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, L., He, XH., Yu, HX. et al. Evaluation of the genetic diversity of mango (Mangifera indica L.) seedling germplasm resources and their potential parents with start codon targeted (SCoT) markers. Genet Resour Crop Evol 67, 41–58 (2020). https://doi.org/10.1007/s10722-019-00865-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-019-00865-8