Abstract
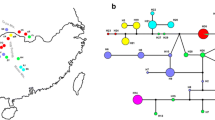
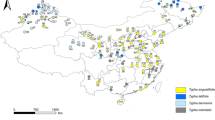
Chinese Cherry, generally referring to Prunus pseudocerasus Lindl. species, is one of the most economically important domestic fruit tree in China. To effectively preserve and sustainably utilize this species, a study in genetic diversity and population structure was carried out by the sequence variation of chloroplast DNA trnQ-rps16 intergenic spacers. Polymorphism was calculated among the 200 individuals from 7 local and 10 wild populations across its geographical range in China. The results showed that (1) Total genetic variation in species level was poor (h = 0.478, π = 0.0018). But in the wild populations, it was much higher than that in the local populations (h = 0.565, π = 0.0021 vs. h = 0.152, π = 0.0006). (2) Low levels of genetic differentiation (FSC = 0.19595, P = 0.000), genetic distance (0.00000–0.00297), as well as pairwise Fst value among populations were detected. These results, combined with the insignificant gene genealogy pattern and neutrality tests indicate that Chinese Cherry has relatively low levels of genetic diversity and insignificant population structure. Coancestry might be a combined effect of the reduction in effective population size with the recent demographic bottlenecks and mating systems. The pronounced seed dispersal abilities and relatively high rates of gene exchange are supported by the limited genetic differentiation among populations.





Similar content being viewed by others
References
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Bio Evol 16:37–48
Birky CWJR (1988) Evolution and variation in plant chloroplast and mitochondrial genomes. In: Gottlieb LD, Jain SK (eds) Plant evolutionary biology. Chapman and Hall, London, pp 23–53
Cai YL, Li S, Cao DW, Qian ZQ, Zhao GF, Han MY (2006) Use of amplified DNA sequences for the genetic analysis of the cherry germplasm. Acta Horticulturae Sinica 33:249–254
Caiedo AL, Schaal BA (2004) Population structure and phylogeography of Solanum pimpinellifolium inferred from a nuclear gene. Mol Ecol 15:1871–1882
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescence theory with applications to intraspecific phylogeny reconstruction. Genetics 134:959–969
Devos N, Tyteca D, Raspé O, Wesselingh RA, Jacquemart AL (2003) Pattern of chloroplast diversity among western European Dactylorhiza species (Orchidaceae). Plant Syst Evol 243:85–97
Donnelly P, Tavare S (1986) The ages of alleles and a coalescent. Adv Appl Probab 18:1–19
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online l:47–50
Forster P, Bandelt HJ, Röhl A (2004) Network 4.2.0.1. Software available free at www.fluxus-engineering.com. Fluxus Technology Ltda
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Fujii T, Nishida M (1997) High sequence variability in the mitochondrial DNA control region of the Japanese flounder Paralichthys olivaceus. Fisheries Sci 63:906–910
Glémin S, Bazin E, Charlesworth D (2006) Impact of mating systems on patterns of sequence polymorphism in flowering plants. P R Soc B-Biol Sci 273:3011–3019
Huang XJ, Chen T, Liang QB, Chen J, Wang XR, Tang HR (2011) Preliminary evaluation on the economic characters of 14 cherry germplasms. South China Fruits 40:15–18
Huang XJ, Wang XR, Chen T, Chen J, Tang HR (2012) Research progress of genetic diversity in Cerasus pseudocerasus and their wild relative populations, and utilize progress of cultivation resources. J Fruit Sci (In press)
Hurka H, Friesen N, German DA, Franzke A, Nenffer B (2012) ‘Missing link’ species Capsella orientalis and Capsella thracica elucidate evolution of model plant genus Capsella (Brassicaceae). Mol Ecol 21:1223–1238
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, NewYork, pp 21–132
Li MM, Cai YL, Qian ZQ, Zhao GF (2009) Genetic diversity and differentiation in Chinese sour cherry Prunus pseudocerasus Lindl., and its implications for conservation. Genet Resour Crop Evol 56:455–464
Librado P, Rozas J (2009) DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu CJ, Liu MZ (1993) The seed identification of the relics from HouMa copper casting site Shaanxi. (Shaanxi Province archaeology institute) HouMa copper casting sites. Cultural relic press, Beijing
Luan SS, Chiang TY, Xun G (2006) High genetic diversity vs. low genetic differentiation in Nouelia insignis (Asteraceae), a narrowly distributed and endemic species in China, revealed by ISSR finger printing. Ann Bot 98:583–589
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Tajima F (1983) Maximum likelihood estimation of the number of nucleotide substitutions from restriction sites data. Genetics 105:207–217
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol 4:406–425
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474
Shaw J, Lickey BE, Schilling EE, Small LR (2007) Comparison of whole chloroplast genome sequences to choose non-coding regions for phylogenetic studies in angiosperms: the Tortoise and the Hare. Ame J Bot 94:275–288
Song CM, Wen XP, Yang ET (2011) Cherry germplasm from Guizhou Province analyzed by ISSR Markers. Acta Horticulturae Sinica 38:1531–1538
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Bio Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Wang FY, Gong X, Hu CM, Hap G (2008) Phylogeography of an alpine species Primula secundiflora inferred from the chloroplast DNA sequence variation. J Syst Evol 46:l3–l22
Wang LY, Abbott RJ, Zheng W, Chen P, Wang YJ, Liu JQ (2009) History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Mol Ecol l8:709–721
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evol 19:395–420
Xun G, Luan SS, Hung KH, Hwang CC, Lin CJ, Chiang YC, Chiang TY (2011) Population structure of Nouelia insignis (Asteraceae), an endangered species in southwestern China, based on chloroplast DNA sequences: recent demographic shrinking. J Plant Res 124:221–230
Yü DJ, Li CL (1986) Flora of China, vol 38. Science Press, Beijing
Yu Y, Downie SR, He X, Deng X, Yan L (2011) Phylogeny and biogeography of Chinese Heracleum (Apiaceae tribe Tordylieae) with comments on their fruit morphology. Plant Syst Evol 296:179–203
Yuan QJ, Zhang ZY, Peng H, Ge S (2008) Chloroplast phylogeography of Dipentodon (Dipentodontaceae) in southwest China and northern Vietnam. Mol Ecol 17:1054–1065
Acknowledgments
We thank the National Natural Science Foundation of China (30971990) and the Key Cultivation Special Project Foundation of Education Department of Sichuan Province (2011A005) for the grant.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, T., Wang, Xr., Tang, Hr. et al. Genetic diversity and population structure of Chinese Cherry revealed by chloroplast DNA trnQ-rps16 intergenic spacers variation. Genet Resour Crop Evol 60, 1859–1871 (2013). https://doi.org/10.1007/s10722-013-9960-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-013-9960-9