Abstract
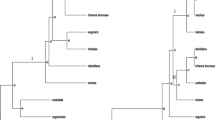
PCR-RFLP analyses of three regions for each of chloroplast DNA (cpDNA; rbcL-ORF106, trnD-trnT, trnH-trnK) and mitochondrial DNA (mtDNA; nad7/exon2-exon3, nad7/exon3-exon4, 18S-5S) were performed in 26 cultivars of acid citrus grown in Japan to identify polymorphisms and classify them. The polymorphisms were compared with those of three true Citrus species, i.e., mandarin, pummelo and citron. Ichang papeda (C. ichangensis) was also included in this study to find its relationship with Yuzu. Inter-species cpDNA variation was recognized and the acid citrus were divided into three groups, namely; I (‘Yuzukichi’ and ‘Kinkoyu’), II [sour oranges (‘Kaiseito’, ‘Daidai’ and ‘China daidai’), ‘Nansho daidai’, ‘Kiku daidai’, C. sudachi (‘Mushi yukaku’, ‘Yushi yukaku’ and ‘Yushi mukaku’), C. sphaerocarpa (‘Kabosu’ and ‘Aka kabosu’), C. kizu (‘Taninaka kizu’, ‘Kinosu’ and ‘Kizu’), ‘Zanbo’, ‘Mochiyu’, ‘Jabara’ and ‘Naoshichi’], and III [Yuzu (‘Tetraploid’, ‘Tochikei yuzu’ and ‘Yamanekei yuzu’), ‘Matsuda sudachi’, ‘Zuishoyu’, ‘Hanayu’ and ‘Yuko’]. CpDNA restriction patterns of the three true Citrus species differed from each other as well as from those of ichang papeda. CpDNA restriction patterns of group I of the acid citrus were identical to those of mandarins. Group II showed the same as pummelos. CpDNA restriction patterns of group III were differed from those of the three true Citrus species in the three regions. This group was differed from ichang papeda after digestion of trnH-trnK PCR products with TaqI, HinfI and AluI, while they showed identical restriction patterns in two regions, rbcL-ORF106 and trnD-trnT. Citrons and ichang papeda were placed in groups IV and V, respectively. Based on mtDNA restriction patterns, the acid citrus were divided into three groups; i, ii and iii. In groups i and ii accessions of groups I and II of cpDNA were placed with mandarins and pummelos, respectively. In group iii accessions of group III of cpDNA were placed with ichang papeda. Citrons were placed in a distinct group, iv.
Similar content being viewed by others
References
Al-Janabi SM, McClelland M, Peterson C, Sorbal BWS (1994) Phylogenetic analysis of organellar DNA sequences in the Andropogoneae: Saccharinea. Theor Appl Genet 88:933–944
Arnold ML, Buckner CM, Robinson JJ (1991) Pollen-mediated introgression and hybrid speciation in Louisiana irises. Proc Nat Acad Sci USA 88:1398–1402
Asadi Abkenar A, Isshiki S (2003) Molecular characterization and genetic diversity among Japanese acid citrus (Citrus spp.) based on RAPD markers. J Hortic Sci Biotechnol 78:108–112
Asadi Abkenar A, Isshiki S, Tashiro Y (2004) Phylogenetic relationships in the “true citrus fruit trees” revealed by PCR-RFLP analysis of cpDNA. Sci Hortic 102:233–242
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131
Dumolin-Lapegue S, Pemonge MH, Petit RJ (1997) An enlarged set of consensus primers for the study of organelle DNA in plants. Mol Ecol 6:393–397
Green RM, Vardi A, Galun E (1986) The plastome of Citrus. Physical map, variation among Citrus cultivars and species and comparison with related genera. Theor Appl Genet 72:170–177
Gulsen O, Roose ML (2001) Chloroplast and nuclear genome analysis of the parentage of lemons. J Am Soc Hort Sci 126:210–215
Hirai M, Kozaki I, Kajiura I (1986) Isozyme analysis and phylogenic relationship of citrus. Japan J Breed 36:377–389
Kimura K, Taninaka T (1988) Acid citrus in Shikoku island. Harada Co., Tokushima, Japan (in Japanese)
Kimura K, Taninaka T (1990) Acid citrus in Shikoku and Kyushu island. Harada Co., Tokushima, Japan (in Japanese)
Kimura K, Taninaka T (1995) Acid citrus in Japan. Harada Co., Tokushima, Japan (in Japanese)
Moreira CD, Gmitter FG Jr, Grosser JW, Huang S, Ortega VM, Chase CD (2002) Inheritance of organelle DNA sequences in a Citrus-Poncirus intergeneric cross. J Hered 93:174–178
Nicolosi E, Deng ZN, Gentile A, La Malfa S, Continella G, Tribulato E (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet 100:1155–1166
Rahman MM, Nito N, Isshiki S (2001) Cultivar identification of ‘Yuzu’ (Citrus junos Sieb. ex Tanaka) and related acid citrus by leaf isozymes. Sci Hortic 87:191–198
Swingle WT, Reece PC (1967) The botany of citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD (eds) The citrus industry, vol 1. Univ Calif Press, Berkeley Calif, USA, pp 389–390
Tanaka T (1954) Species problem in citrus. Jpn Soc for the Promotion of Science, Tokyo, p 152
Tanaka T (1977) Fundamental discussion of Citrus classification. Stud Citrol 14:1–6
Taninaka T, Otoi N, Morimoto J (1981). Acid citrus cultivars related to the Yuzu (Citrus junos Sieb. ex Tanaka) in Japan. In: Matsumoto K, Oogaki C, Kozaki I (eds) Proc Int Soc Citriculture. Aiko Printing Co., Tokyo, Japan, pp 73–76
Yamamoto M, Kobayashi S, Nakamura Y, Yamada Y (1993) Phylogenic relationships of citrus revealed by RFLP analysis of mitochondrial and chloroplast DNA. Japan J Breed 43:355–365
Yamamoto M, Kobayashi S (1996) Polymorphism of chloroplast DNA in citrus. J Japan Soc Hort Sci 65:291–296
Zhusheng C (1997) Citrus germplasm in China. In: Sykes SR, Bevington KB, Hailstones D (eds) Proceedings citrus germplasm conservation workshop. Brisbane, Australia, pp 85–95
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abkenar, A.A., Isshiki, S., Matsumoto, R. et al. Comparative analysis of organelle DNAs in acid citrus grown in Japan using PCR-RFLP method. Genet Resour Crop Evol 55, 487–492 (2008). https://doi.org/10.1007/s10722-007-9253-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-007-9253-2