Abstract
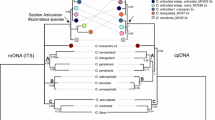
To study the phylogenetics of sugarcane (Saccharum officinarum L.) and its relatives we sequenced four loci on cytoplasmic genomes (two chloroplast and two mitochondrial) and analyzed mitochondrial RFLPs generated using probes for COXI, COXII, COXIII, Cob, 18S+5S, 26S, ATPase 6, ATPase 9, and ATPase α (D'Hont et al. 1993). Approximately 650 bp of DNA in the intergenic spacer region between rbcL and atpB and approximately 150 bp from the chloroplast 16S rDNA through the intergenic spacer region tRNAval gene were sequenced. In the mitochondrial genome, part of the 18S rRNA gene and approximately 150 bp from the 18S gene 3′ end, through an intergenic spacer region, to the 5S rRNA gene were sequenced. No polymorphisms were observed between maize, sorghum, and ‘Saccharum complex’ members for the mitochondrial 18S internal region or for the intergenic tRNAval chloroplast locus. Two polymorphisms (insertion-deletion events, indels) were observed within the 18S-5S mitochondrial locus, which separated the accessions into three groups: one containing all of the Erianthus, Eccoilopus, Imperata, Sorghum, and 1 Miscanthus species; a second containing Saccharum species, Narenga porphyrocoma, Sclerostachya fusca, and 1 presumably hybrid Miscanthus sp. from New Guinea; and a third containing maize. Eighteen accessions were sequenced for the intergenic region between rbcL and atpB, which was the most polymorphic of the regions studied and contained 52 site mutations and 52 indels, across all taxa. Within the Saccharum complex, at most 7 site mutations and 16 indels were informative. The maternal lineage of Erianthus/Eccoilopus was nearly as divergent from the remaining Saccharum complex members as it was from sorghum, in agreement with a previous study. Sequences from the rbcL-atpB spacer were aligned with GENBANK sequences for wheat, rice, barley, and maize, which were used as outgroups in phylogenetic analyses. To determine whether limited intra-complex variability was caused by under sampling of taxa, we used seven restriction enzymes to digest the PCR-amplified rbcL-atpB spacer of an additional 36 accessions within the Saccharum complex. This analysis revealed ten restriction sites (none informative) and eight length variants (four informative). The small amount of variation present in the organellar DNAs of this polyploid complex suggests that either the complex is very young or that rates of evolution between the Saccharum complex and outgroup taxa are different. Other phylogenetic information will be required to resolve systematic relationships within the complex. Finally, no variation was observed in commercial sugarcane varieties, implying a world-wide cytoplasmic monoculture for this crop.
Similar content being viewed by others
References
Albert VA, Mischler BD, Chase MW (1992) Character-state weighting for restriction site data in phylogenetic reconstruction, with an example from chloroplast DNA. In: Soltis PS, Soltis DE, and Doyle JJ (eds) Molecular systematics of plants, Chapman and Hall, New York, pp 349–403
Al-Janabi SM, Honeycutt RJ, McClelland M, Sobral BWS. (1993) A genetic linkage map of Saccharum spontaneum L. ‘SES 208’. Genetics 134:1249–1260
Artschwager E (1954) A taxonomic study of Saccharum sinense Roxb. and S. barberi Jeswiet. USDA Technical Bulletin No. 1089, Washington D.C.
Artschwager E, Brandes EW (1958) Sugarcane (Saccharum officinarum L.): origin, classification, characteristics, and descriptions of representative clones. USDA Handbook No. 122, Washington, D. C.
Bellwood P (1985) Prehistory of the Indo-Malaysian archipelago. Academic Press, Orlando, Fa.
Birky CW (1988) Evolution and variation in plant chloroplast and mitochondrial genomes. In Gottlieb LD, Iain SK (eds): “Plant evolutionary biology”. Chapman and Hall, London, pp. 23–53
Bowman CM, Bonnard G, Dyer TA (1983) Chloroplast DNA variation between species of Triticum and Aegilops. Location of the variation on the chloroplast genome and its relevance to the inheritance and classification of the cytoplasm. Theor Appl Genet 65:247–262
Brandes EW (1928) Into primeval Papua by seaplane. Natl Geographic 56:253–332
Brandes EW, Sartoris GB, Grassl CO (1939) Assembling and evaluating wild forms of sugarcane and closely related plants. In: Proc. 6th Int Soc Sugarcane Technol Congr., pp. 128–154
Bremer B (1991) Restriction site data from chloroplast DNA for phylogenetic reconstruction: is there only one accurate way of scoring? Plant Syst Evol 175:39–54
Burner DM (1991) Cytogenetic analyses of sugarcane relatives in the subtribe Saccharinae. Euphytica 54:125–133
Celarier RP (1956) Cytotaxonomy of the Andropogoneae. 1. Subtribes Dimeriinae and Saccharinae. Cytologia 21:272–291
Clayton WD, Renvoize SA (1986) Genera Graminum. Grasses of the world. Kew Bulletin Additional Series XIII, Royal Botanical Gardens, Kew, Her Majesty's Stationery Office, London
Da Silva J, Sorrells ME, Burnquist WL, Tanksley SD (1993) RFLP linkage map and genome analysis of Saccharum spontaneum. Genome, in press
Daniels J, Roach BT (1987) Taxonomy and evolution in breeding. In: Heinz DJ (ed). Sugarcane improvement through breeding. Elsevier, Amsterdam, pp 7–84
Daniels J, Williams CA (1975) The origin of the genus Saccharum. ISSCT Sugarcane Breed Newsl 36:24–39
Daniels J, Paton H, Smith P, Williams C (1980) Further studies on leaf flavonoids as evolutionary indicators in Saccharum officinarum L. In: Proc 17th XVII ISSCT Congr., pp. 1317–1335
D'Hont A, Lu YH, Feldmann P, Glaszmann JC (1993) Cytoplasmic diversity in sugarcane revealed by heterologous probes. Sugar Cane 1993:12–15
Doebley J, Stec A (1991) Genetic analysis of the morphological differences between maize and teosinte. Genetics 129:285–295
Duvall MR, Doebley J (1990) Restriction site variation in the chloroplast genome of Sorghum (Poaceae). Syst Bot 15:472–480
Felsenstein J (1989) PHYLIP- phylogeny inference package. Cladistics 5:164–166
Glaszmann JC, Lu YH, Lanaud C (1990). Variation of nuclear ribosomal DNA in sugarcane. J Genet Breed 44:191–198
Hamby RK, Zimmer EA (1988) Ribosomal RNA sequences for inferring phylogeny within the grass family (Poaceae). Plant Syst Evol 160:29–37
Hartley W (1958) Studies on the origin, evolution, and distribution of the Graminiae. I. The tribe Andropogoneae. Aust J Bot 6:116–128
Honeycutt RJ, Sobral BWS, Keim P, Irvine JE (1992) A rapid DNA extraction method for sugarcane and its relatives. Plant Mol Bio Rep 10:66–72
Liston A (1992) Variation in the chloroplast genes rpoc1 and rpoc2 of the genus Astragalus (Fabaceae): evidence from restriction site mapping of a PCR-amplified fragment. Am J Bot 79:953–961
Maniatis T, Fritch EF, Sambrook J (1982) Molecular cloning labrotary manual. Cold Spring Harbor Labrotary Press, Cold Spring Harbor, N.Y.
McIntosh L, Poulsen C, Bogorad L (1980) Chloroplast gene sequence for the large subunit of ribulose bisphosphatecarboxylase of maize. Nature 288:556–560
Mohan N, Sreenivasan TV (1983) Chromosome number in the genus Erianthus, Michx. (Poaceae) of Indonesian Arquipelago. Cell Chromosome Res 6:14–16
Moriya A (1940) List of chromosome numbers in the genus Saccharum and related genera. Jpn J Genet 16:126–136
Mukherjee SK (1957) Origin and distribution of Saccharum. Bot Gaz 119:55–61
Panje RR, Babu CN (1960) Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia 25:152–172
Price S (1957) Cytological studies in Saccharum and allied genera. III. Chromosome numbers in interspecific hybrids. Bot Gaz (March): 146–159
Price S (1968) Chromosome transmission by Saccharum robustum in interspecific crosses. J Hered 59:245–247
Ralph D McClleland M Welsh J, Branton G, Perolat P (1993) Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J Bacteriol 175:973–981
Ruanto G, Kidd KK (1991) Coupled amplification and sequencing of genomic DNA. Proc Natl Acad Sci USA 88:2815–2819
Salts Y, Herrmann RG, Peleg N, Lavi U, Izhar S, Frankel R, Beckmann JS (1984) Physical mapping of plastid DNA variation among eleven Nicotiana species. Theor Appl Genet 69:1–14
Sanger F, Coulsen AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94:441–448
Smith RM, Martin-Smith M (1978) Triterpene methyl ethers in leaf waxes of Saccharum and related genera. Phytochemistry 17:1307–1312
Sobral BWS, Braga DPV, LaHood E, Keim P (1994) Phylogenetic analysis of chloroplast restriction enzyme site mutations in the Saccharinae: Andropogoneae. Theor Appl Genet (in press)
Soltis PS, Doyle JJ, Soltis DE (1992) Molecular data and polyploid evolution in plants. In: Soltis PS, Soltis DE, Doyle JJ (eds). Molecular systematics of plants. Chapman and Hall, New York, pp 177–201
Springer PS, Zimmer EA, Bennetzen JL (1989) Genomic organization of the ribosomal DNA of sorghum and its close relatives. Theor Appl Genet 77:844–850
Stebbins GL (1956) Cytogenetics and evolution of the grass family. Am J Bot 43:890–905
Swofford DL (1991) PAUP: Phylogenetic Analysis Using Parsimony, version 3.1 s. Computer program distributed by the Illinois Natural History Survey, Champaign, Ill.
Swofford DL, Olsen GJ (1990) Phylogeny reconstruction. In: Hillis DM, Moritz C (eds) Molecular Systematics. Sinauer Assoc, Sunderland, Mass., pp. 411–501
Waldron JC, Glasziou KT, Daniels J (1974) B-amylase isoenzymes as genetic markers in Saccharum and related genera. In: Proc. 15th ISSCT Congr. pp. 145–152
Watson L, Clifford HT, Dallwitz MJ (1985) The classification of the Poaceae: subfamilies and supertribes. Aust J Bot 33:433–484
Williams CA, Harborne JB (1974) The taxonomic significance of leaf flavonoids in Saccharum and related genera. Phytochemistry 13:1141–1149
Wilson MA, Gaut B, Clegg MT (1990) Chloroplast DNA evolves slowly in the palm family (Arecaceae). Mol Biol Evol 7:303–314
Zurawski G, Clegg MT (1987) Evolution of higher-plant chloroplast DNA-encoded genes: implications for structure-function and phylogenetic studies. Annu Rev Plant Physiol 38:391–418
Zurawski G, Clegg MT, Brown AHD (1984) The nature of nucleotide sequence divergence between barley and maize chloroplast DNA. Genetics 106:735–749
Author information
Authors and Affiliations
Additional information
Communicated by A. R. Hallauer
Rights and permissions
About this article
Cite this article
Al-Janabi, S.M., McClelland, M., Petersen, C. et al. Phylogenetic analysis of organellar DNA sequences in the Andropogoneae: Saccharinae. Theoret. Appl. Genetics 88, 933–944 (1994). https://doi.org/10.1007/BF00220799
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00220799