Abstract
The Emilia region (Northern Italy) is characterised by the occurrence of microclimates that permit olive growing. The presence of the species, albeit sporadic, in these territories for several centuries as a fruit crop is well documented, by both archaeological and written testimony, and by a large number of plants well over a century old, located in particular sites, favourable for growth and development of the tree.
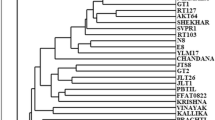
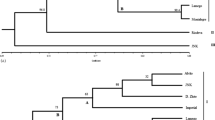
Olive genetic diversity was studied using RAPD and SSR techniques, on plants growing in the Emilia territory (Reggio Emilia and Parma provinces). For genotype identification comparisons were made with 8 cultivars, some of which from Central Italy. Screening was obtained analysing patterns produced by 20 RAPD primers and 3 SSR primers, developed by other authors; the primers and we were able to discriminate olive cultivars with a sufficient degree of reliability. The dendrograms obtained from the analysis show the genetic relationship among accessions present in the Parma-Reggio Emilia district. Our results demonstrated the reliability of RAPDs and SSRs to identify all studied olive cultivars and to reveal the degree of their relatedness to each other. The analysis also reveals the presence of an interesting amount of genetic diversity among the studied individuals.



Similar content being viewed by others
References
Amane M, Lumeret R, Hany V, Ouazzani N, Debain C, Vivier G, Deguilloux MF (1999) Chloroplast-DNA variation in cultivated and wild olive (Olea europaea L.). Theor Appl Genet 99:133–139
Angiolillo A, Mencuccini M, Baldoni L (1999) Olive genetic diversity assessed using amplified fragment length polymorphism. Theor Appl Genet 98:411–421
Barranco D, Cimato A, Fiorino P, Rallo L, Touzani A, Castaneda C, Serafin F, Trujillo I (2000) World catalogue of olive varieties. International Olive Oil Concil, Madrid, 360 pp
Bartolini G, Prevost G, Messeri C, Carignani G (2005) Olive germplasm: cultivars and word-wide collections. Web site FAO: http://www.apps3.fao.org/wiews/olive/oliv.jsp
Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, Trujillo I (2003) Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor Appl Genet 107:736–744
Belaj A, Satovic Z, Rallo J, Trujillo I (2004a) Optimal use of RAPD markers for identifying varieties in olive (Olea europaea L.) germplasm collection. J Am Soc Hortic Sci 129(2):266–270
Belaj A, Cipriani G, Testolin R, Rallo L, Trujillo I (2004b) Characterization and identification of the main Spanish and Italian olive cultivars by simple-sequence-repeat markers. HortScience 39(7):1557–1561
Besnard G, Khadari B, Baradat P, Bervillé A (2002) Olea europaea (Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theor Appl Genet 104:1353–1361
Besnard G, Bervillé A (2000) Multiple origins for Mediterranean olive (Olea europaea L. ssp. europaea) based upon mitochondria DNA polymorphisms. Life Science 323:173–181. CR Acad Sci Paris
Bianchedi C (1880) L’Olivo sulle colline parmensi. Ferrari e figli, Parma Italy, 60 pp
Bignardi A (1978) Le campagne emiliane nel rinascimento e nell’età barocca. Forni, Bologna Italy, 401 pp
Carriero F, Fontanazza G, Cellini F, Giorio G (2002) Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor Appl Genet 104:301–307
Cimato A, Cantini C, Sani G (2001) L’olivo in Toscana: il germoplasma autoctono. ARSIA Toscana-IPSL, CNR-Regione Toscana, Florence, Italy
Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R (2002) Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor Appl Genet 104:223–228
Diaz Bermudez A (2005) Desarrollo y caracterización de nuevos microsatélites y SNPs y aplicación en la mejora genética del olivo (Olea europaea L.), Ph.D. tesis, Departamento de Genética, Universidad de Córdoba—ETSIAM Córdoba, Spain, 180 pp
Dice LR (1945) Measures of the amount of ecological association between species. Ecology 26:297–302
Fabbri A (2005) Olivicoltura emiliana? Perchè no! Olivo e Olio 4:12–17
Fabbri A, Ganino T (2003) Il germoplasma olivicolo in Emilia. Atti Convegno Germoplasma olivicolo e tipicità dell’olio Perugia, Italy, pp 48–52
Fabbri A, Hormaza JI, Polito VS (1995) Random amplified polymorphic DNA analysis of olive (Olea europaea L.) cultivars. J Am Soc Hort Sci 120:538–542
Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Fragaki G, Spyros A, Siragakis G, Salivaras E, Dais P (2005) Detection of extra vergin olive oil adulteration with lampante olive oil and refined olive oil using nuclear magnetic resonance spectroscopy and multivariate statistical analysis. J Agric Food Chem 53:2810–2816
Gallitelli M, Semeraro L, Antonelli NM (1991) RFLP analysis in the olive (Olea europaea L.). EMBO Course, Cologne
Ganino T, Bartolini A, Fabbri A (2006) The classification of olive germplasm—a review. J Hortic Sci Biotech 81(3):319–334
Ganino T, Fabbri A (2005) Genetic characterization of Olea europaea L. germplasm in northern Italy. 5th international symposium on olive growing, 27 September–2 October 2004, Izmir (Turkey), 127 pp
Gounaris Y, Skoula M, Fournaraki C, Drakakaki G, Makris A (2002) Comparison of essential oils and genetic relationship of Origanum × intercedens to its parental taxa in the island of Crete. Biochem Syst Ecol 30:249–258
Jaccard P (1908) Nouvelle recherches sur la distribution florale. B Soc Vaud Sci Nat 44:223–270
Kirby LT (1990) DNA fingerprinting: an introduction, Stockton Press, New York, 365 pp
Loukas M, Krimbas CB (1983) History of olive cultivars based on their genetic distances. J Hortic Sci 58:121–127
Mancuso S, Nicese FP (1999) Identifying olive (Olea europaea L.) cultivars using Artificial Neural Networks. J Am Soc Hortic Sci 124(5):527–531
Marrazzo T, Cipriani G, Marconi R, Cimato A, Testolin R (2002) Isolation and characterisation of microsatellite DNA in Olive (Olea europaea L.). Acta Hort 586:61–64
Matsuoka Y, Mitchell SE, Kresovich S, Goodman M, Doebley J (2002) Microsatellites in Zea—variability, patterns of mutations, and use for evolutionary studies. Theor Appl Genet 104:436–450
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Owen CA, Bita EC, Banilas G, Hajjar SE, Sellianakis V, Aksoy U, Hepaksoy S, Chamoun R, Talhook SN, Metzidakis I, Hatzopoulos P, Kalaitzis P (2005) AFLP reveals structural details of genetic diversity within cultivated olive germplasm from the Eastern Mediterranean. Theor Appl Genet 110:1169–1176
Ozkaya MT, Cakir E, Gokbayrak Z, Ercan H, Taskin N (2006) Morphological and molecular characterization of Derik Halhali olive (Olea europaea L.) accession grown in Derik-Mardin province of Turkey. Sci Hortic 108:205–209
Peakall R, Smouse PE (2005) GenAlEx 6: Genetic Analysis in Excel. Population genetic software for teaching and research. The Australian National University, Canberra, Australia. http://www.anu.edu.au/BoZo/GenAlEx/
Podani J (2001) Sin-tax 2000 Computer program for data analysis in ecology and systematics: user’s manual. Budapest, Hungary
R Development Core Team (2005) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0, URL http://www.R-project.org
Rallo P, Dorado G, Martin A (2000) Development of simple sequence repeats (SSRs) in olive tree (Olea europaea L.). Theor Appl Genet 101:984–989
Rotondi A, Mari M, Babini AR, Govoni M, Cristoferi G (2004) L’attitudine alla propagazione e la certificazione genetica e sanitaria dell’olivo in Emilia-Romagna. Regione Emilia-Romagna, CNR-IBIMET, Bologna, Italy, 117 pp
Sanz-Cortés F, Parfitt DE, Romero C, Struss D, Liácer G, Badenes M (2003) Intraspecific olive diversity assessed with AFLP. Plant Breed 122:173–177
Sefc KM, Lopes MS, Mendonça D, Rodrigues Dos Santos M, Laimer Da Câmara Machado M, Da Câmara Machado A (2000) Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol Ecol 9:1171–1173
Trujillo I, Rallo L, Arus P (1995). Identifying olive cultivars by isozyme analysis. J Am Soc Hortic Sci 120:318–324
Wagner HW, Sefc KM (1999) Identity 1.0: freeware program for the analysis of microsatellite data. Vienna, http://www.boku.ac.at/zag/forsch/identity.htm
Vlahov G, Shaw AD, Kell DB (1999) Use of 13 C nuclear magnetic resonance distortionless enhancement by polarization transfer pulse sequence and multivariate analysis to discriminate olive oil cultivars. J Am Oil Chem Soc 76(10):1223–1231
Acknowledgements
The Authors wish to thank Dr. Enzo Perri, Director of Istituto Sperimentale per l’Olivicoltura of Rende (Cosenza, Italy), for the germplasm made available for this research, and Dr. Paolo Piovani, for reviewing the manuscript. The research was supported by the Project “Studio sullo sviluppo dell’olivicoltura da olio in Emilia-Romagna, nelle province di Modena, Reggio Emilia, Parma e Piacenza”, made possible by a joint grant of the said Provinces and of the Region Emilia-Romagna.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ganino, T., Beghè, D., Valenti, S. et al. RAPD and SSR markers for characterization and identification of ancient cultivars of Olea europaea L. in the Emilia region, Northern Italy . Genet Resour Crop Evol 54, 1531–1540 (2007). https://doi.org/10.1007/s10722-006-9145-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-006-9145-x