Abstract
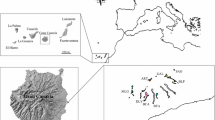
Bethencourtia Choisy ex Link is an endemic genus of the Canary Islands and comprises three species. Bethencourtia hermosae and Bethencourtia rupicola are restricted to La Gomera, while Bethencourtia palmensis is present in Tenerife and La Palma. Despite the morphological differences previously found between the species, there are still taxonomic incongruities in the group, with evident consequences for its monitoring and conservation. The objectives of this study were to define the species differentiation, perform population genetic analysis and propose conservation strategies for Bethencourtia. To achieve these objectives, we characterized 10 polymorphic SSR markers. Eleven natural populations (276 individuals) were analyzed (three for B. hermosae, five for B. rupicola and three for B. palmensis). The results obtained by AMOVA, PCoA and Bayesian analysis on STRUCTURE confirmed the evidence of well-structured groups corresponding to the three species. At the intra-specific level, B. hermosae and B. rupicola did not show a clear population structure, while B. palmensis was aggregated according to island of origin. This is consistent with self-incompatibility in the group and high gene flow within species. Overall, the genetic diversity of the three species was low, with expected heterozygosity values of 0.302 (B. hermosae), 0.382 (B. rupicola) and 0.454 (B. palmensis). Recent bottleneck events and a low number of individuals per population are probably the causes of the low genetic diversity. We consider that they are naturally rare species associated with specific habitats. The results given in this article will provide useful information to assist in conservation genetics programs for this endemic genus.




Similar content being viewed by others
References
Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J (2008) Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188. https://doi.org/10.1111/j.1365-294X.2008.03971.x
Arechavaleta M, Rodríguez S, Zurita N, García A (2010) Lista de especies silvestres de Canarias (hongos, plantas y animales terrestres). Gobierno de Canarias
Barrett S, Kohn J (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 3–30
BOC (2010) Ley 4/2010 de 4 de junio, del Catálogo canario de especies protegidas. Boletín Oficial de Canarias 112:15200–15225
Brennan AC, Barker D, Hiscock SJ, Abbott RJ (2012) Molecular genetic and quantitative trait divergence associated with recent homoploid hybrid speciation: a study of Senecio squalidus (Asteraceae). Heredity 108:87–95. https://doi.org/10.1038/hdy.2011.46
Caujapé-Castells J, Tye A, Crawford DJ, Santos-Guerra A, Sakai A, Beaver K, Lobin W, Vincent Florens FB, Moura M, Jardim R, Gómes I, Kueffer C (2010) Conservation of oceanic island floras: Present and future global challenges. Perspect Plant Ecol Evol Syst 12:107–129. https://doi.org/10.1016/j.ppees.2009.10.001
Cole CT (2003) Genetic variation in rare and common plants. Annu Rev Ecol Evol Syst 34:213–237. https://doi.org/10.2307/30033775
Cornuet JM, Luikart G (1996) Power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crawford DJ, Stuessy TF (2016) Cryptic variation, molecular data, and the challenge of conserving plant diversity in oceanic archipelagos: the critical role of plant systematics. Taxon 46:129–148. https://doi.org/10.11110/kjpt.2016.46.2.129
Crawford DJ, Lowrey TK, Anderson GJ, Bernardello G, Santos-guerra A, Stuessy TF (2009) Genetic diversity in Asteraceae endemic to oceanic islands: Baker’ s Law and polyploidy. In: Funk V, Susanna A, Stuessy TF, Bayer R (eds) Systematic, evolution and biogeography of Compositae. International Association for Plant Taxonomy edn, Vienna, Austria, pp 101–114
David P, Pujol B, Viard F, Castella V, Goudet J (2007) Reliable selfing rate estimates from imperfect population genetic data. Mol Ecol 16:2474–2487. https://doi.org/10.1111/j.1365-294X.2007.03330.x
Dellaporta S, Wood L, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 14:19–21
DeSalle R, Amato G (2004) The expansion of conservation genetics. Nat Rev Genet 5:702–712. https://doi.org/10.1038/nrg1425
Dias EF, Sardos J, Silva L, Maciel MGB, Moura M (2014) Microsatellite markers unravel the population genetic structure of the Azorean Leontodon: Implications in conservation. Plant Syst Evol 300:987–1001. https://doi.org/10.1007/s00606-013-0937-0
Earl DA, vonHoldt BM (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–242. https://doi.org/10.1146/annurev.ecolsys.24.1.217
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fernández-López ÁB, Gómez-González LAG, Gómez M (2014) Garajonay después del gran incendio de 2012. In: Santamarta Cerezal JC (ed) Investigación, gestión y técnica forestal, en la región de la Macaronesia. Colegio de Ingenieros de Montes, Madrid, pp 201–226
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993. https://doi.org/10.1534/genetics.108.092221
Francisco-Ortega J, Santos-Guerra A, Kim SC, Crawford DJ (2000) Plant genetic diversity in the Canary Islands: a conservation perspective. Am J Bot 87:909–919
Frankham R (1997) Do island populations have less genetic variation than mainland populations?. Heredity 78(Pt 3):311–327
Frankham R (1998) Inbreeding and extinction: Island populations. Conserv Biol 12:665–675
Frankham R, Briscoe D, Ballou J, Briscoe D (2002) Introduction to Conservation Genetics. Cambridge University Press, Cambridge
García-Verdugo C, Baldwin BG, Fay MF, Caujapé-Castells J (2014) Life history traits and patterns of diversification in oceanic archipelagos: A meta-analysis. Bot J Linn Soc 174:334–348
Gobierno de Canarias (2009) Evaluación de especies catalogadas de Canarias. Senecio hermosae. Available at: http://www.gobiernodecanarias.org/opencmsweb/export/sites/medioambiente/piac/galerias/descargas/Documentos/Biodiversidad/evaluacion-especies/evaluacion-2009/Senecio_hermosae_09.pdf. Accessed October 2016
González-Pérez M, Caujapé-Castells J (2014) Development and characterization of nuclear microsatellite markers for Parolinia ornata Webb (Brassicaceae), and cross-species amplification in all species described in the Canarian endemic genus Parolinia. Conserv Genet Resour 6:705–706
González-Pérez M, Lledó MD, Lexer C, Fay M, Marrero M, Bañares-Baudet Á, Carqué E, Sosa PA (2009a) Genetic diversity and differentiation in natural and reintroduced populations of Bencomia exstipulata and comparisons with B. caudata (Rosaceae) in the Canary Islands: an analysis using microsatellites. Bot J Linn Soc 160:429–441. https://doi.org/10.1111/j.1095-8339.2009.00986.x
González-Pérez M, Sosa PA, Rivero E, González-González EA, Naranjo A (2009b) Molecular markers reveal no genetic differentiation between Myrica rivas-martinezii. and M. faya (Myricaceae). Ann Bot 103:79–86. https://doi.org/10.1093/aob/mcn222
Hardy OJ, Vekemans X (2002) SPAGeDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
IUCN (2012) IUCN red list categories and criteria: Version 3.1. IUCN, Gland
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Jarne P, Charlesworth D (1993) Selfing rate in functionally hermaphrodite plants. Annu Rev Ecol Evol Syst 24:441–466
Jeffrey C (1992) The tribe Senecioneae (Compositae) in the Mascarene Islands with an Annotated World Check-List of the genera of the tribe: notes on Compositae : VI. Kew Bull 47:49–109
Kalinowski ST (2005) HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Kreft H, Jetz W, Mutke J, Kier G, Barthlott W (2008) Global diversity of island floras from a macroecological perspective. Ecol Lett 11:116–127. https://doi.org/10.1111/j.1461-0248.2007.01129.x
Kunkel G (1975) Novedades y Taxones Críticos en la Flora de La Gomera. Cuad Bot Canar 25:17–49
Langella O (2002) POPULATIONS 1.2. 28. Population genetic software (individuals or populations distances, phylogenetic trees). CNRS, France. http://bioinformatics.org/~tryphon/populations/. Accessed 28 Mar 2016
Mairal M, Sanmartín I, Aldasoro JJ, Culshaw V, Manolopoulou I, Alarcón M (2015) Palaeo-islands as refugia and sources of genetic diversity within volcanic archipelagos: The case of the widespread endemic Canarina canariensis (Campanulaceae). Mol Ecol 24:3944–3963. https://doi.org/10.1111/mec.13282
Martín Osorio VE, Wildpret de la Torre W, Alcántara Vernet E (2011) Canariothamnus hermosae. The IUCN Red List of Threatened Species 2011: e.T195482A8972929. http://dx.doi.org/10.2305/IUCN.UK.2011-1.RLTS.T195482A8972929.en. Accessed 23 Feb 2017
Médail F, Quézel P (1997) Hot-spots analysis for conservation of plant biodiversity in the Mediterranean Basin. Ann Missouri Bot Gard 84:112. https://doi.org/10.2307/2399957
Meloni M, Reid A, Caujapé-Castells J, Soto M, Fernández-Palacios JM, Conti E (2015) High genetic diversity and population structure in the endangered Canarian endemic Ruta oreojasme (Rutaceae). Genetica 143:571–580. https://doi.org/10.1007/s10709-015-9855-0
Moreno-Saiz JC (2010) Lista Roja de la Flora Vascular Española. Madrid: Dirección General de Medio Natural y Política Forestal (Ministerio de Medio Ambiente, y Medio Rural y Marino)-Sociedad Española de Biología de la Conservación de Plantas
Moreno-Saiz JC, Domínguez-Lozano F, Marrero-Gómez M, Bañares-Baudet Á (2015) Application of the Red List Index for conservation assessment of Spanish vascular plants. Conserv Biol 29:910–919. https://doi.org/10.1111/cobi.12437
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol 19:153–170. https://doi.org/10.1007/BF02300753
Nordenstam B (2006a) Canariothamnus B. Nord., a new genus of the Compositae-Senecioneae, endemic to the Canary Islands. Compos Newslett 44:24–31
Nordenstam B (2006b) Bethencourtia. In: Greuter W, Raab-straube E (eds) Wildenowia vol 2. Notulae adfloram Euro-Mediterranean Pertinentes, pp 707–717
Nordenstam B, Pelser PB, Kadereit JW, Watson LE (2009) Senecioneae. In: Funk V, Susanna A, Stuessy T, Bayer R (eds) Systematics, evolution, and biogeography of the Compositae. International Association for Plant Taxonomy edn. Vienna,, Austria, pp 503–525
Ortega C, González C (1986) Contribución a la conservación “ex sito” de especies canarias en peligro: Propagación “in vitro” de Senecio hermosae Pitard. Bot macaronesica 14:59–72
Peakall R, Smouse PE (2012) GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pelser PB, Nordenstam B, Kadereit JW (2007) An ITS Phylogeny of tribe Senecioneae (Asteraceae) and a New Delimitation of Senecio L. Taxon 56:1077–1104
Pérez de Paz JP, Caujapé-Castells J (2013) A review of the allozyme data set for the Canarian endemic flora: causes of the high genetic diversity levels and implications for conservation. Ann Bot 111:1059–1073. https://doi.org/10.1093/aob/mct076
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a program for detecting recent effective population size reductions from allele data frequencies. J Hered 90:502–503
Pritchard JK, Stephens MM, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1111/j.1471-8286.2007.01758.x
Rambaut A (2009) FigTree v1. 3.1: Tree figure drawing tool. In: http://tree.bio.ed.ac.uk/software/figtree. Accessed October 2016
Reyes-Betancort JA, Santos Guerra A, Guma IR, Humphries CJ, Carine MA (2008) Diversity, rarity and the evolution and conservation of the Canary Islands endemic flora. An del Jardín Botánico Madrid 65:25–45. https://doi.org/10.3989/ajbm.2008.v65.i1.244
Rice W (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rousset F (2008) GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Silva LB, Sardos J, de Sequeira MM, Silva L, Crawford D, Moura M (2016) Understanding intra and inter-archipelago population genetic patterns within a recently evolved insular endemic lineage. Plant Syst Evol 302:367–384. https://doi.org/10.1007/s00606-015-1267-1
Sosa PA, González-Pérez M, Moreno C, Clarke JB (2010) Conservation genetics of the endangered endemic Sambucus palmensis Link (Sambucaceae) from the Canary Islands. Conserv Genet 11:2357–2368. https://doi.org/10.1007/s10592-010-0122-8
Sosa PA, González-Pérez M, González-González EA, Rivero E (2011) Genetic diversity of Canarian endemisms revealed by microsatellites: knowledge after one decade of analysis. In: Caujapé-Castells J, Nieto Feliner G, Fernández-Palacios JM (eds) Proceedings of the Amurga International Conferences on Island Biodiversity. Maspalomas, pp 94–100
Sosa PA, González-González EA, González-Pérez M, Pérez de Paz PL (2013) Contrasting patterns of genetic differentiation in Macaronesian lineages of Ilex (Aquifoliaceae). Bot J Linn Soc 173:258–268. https://doi.org/10.1111/boj.12067
Soto ME, Marrero Á, Roca-Salinas A, Bramwell D, Caujapé-Castells J (2016) Conservation implications of high genetic variation in two closely related and highly threatened species of Crambe (Brassicaceae) endemic to the island of Gran Canaria: C. tamadabensis and C. pritzelii. Bot J Linn Soc 182:152–168. https://doi.org/10.1111/boj.12463
Van Oosterhout C, Weetman D, Hutchinson WF (2006) Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol Ecol Notes 6:255–256. https://doi.org/10.1111/j.1471-8286.2005.01082.x
Vitales D, García-Fernández A, Pellicer J, Vallès J, Santos-Guerra A, Cowan RS, Fay MF, Hidalgo O, Garnatje T (2014) Key processes for Cheirolophus (Asteraceae) diversification on Oceanic Islands inferred from AFLP data. PLoS One 9:1–14. https://doi.org/10.1371/journal.pone.0113207
Weir BS, Cockerham CC (1984) Estimating F-Statistics for the Analysis of Population Structure. Evolution 38:1358–1370
Whittaker RJ, Fernández-Palacios JM (2007) Island biogeography. ecology, evolution and conservation, Second Edi. Oxford University Press, Oxford and La Laguna
Acknowledgements
This research was funded by “Organismo Autónomo de Parques Nacionales” (Project 255/2011). We thank ‘‘Agencia Canaria de Investigación, Innovación y Sociedad de la Información’’ for the fellowship granted to Priscila Rodríguez. We are also grateful to Ángel Fernández, Sito Chinea and Ángel García from Garajonay National Park and the colleagues Pedro Romero, Agustín Naranjo Cigala, Claudio Moreno Medina, Juan José Robledo Arnuncio, Miguel Ángel González and Leticia Curbelo for the help in the samples collection and the laboratory work. Montserrat Fernández and Isabel Saro have added useful comments to the final version.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rodríguez-Rodríguez, P., Pérez de Paz, P.L. & Sosa, P.A. Species delimitation and conservation genetics of the Canarian endemic Bethencourtia (Asteraceae). Genetica 146, 199–210 (2018). https://doi.org/10.1007/s10709-018-0013-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-018-0013-3