Abstract
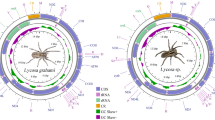
The complete mitogenome of the jumping spider Carrhotus xanthogramma was determined and comparative analysis among four salticid mitogenomes was conducted. The circular genome is 14,563 bp in size and contains a complete set of genes that usually present in the metazoa. All of the 13 protein-coding genes begin with a typical ATN codon and stop with the canonical stop codons, except for ND4 and ND4L genes with an incomplete stop codon T. All of the tRNAs cannot be formed the fully paired acceptor stems and seven out of them cannot be folded into the typical cloverleaf-shaped secondary structures. The tRNA Glu gene translocates its position as compared to the mitogenomes of other three determined jumping spiders. The A+T content of the majority strand and the A+T-rich region are 75.1 and 80%, respectively. The phylogenetic relationships based on concatenated nucleotide and amino acid sequences of 13 protein-coding genes using Maximum Likelihood and Bayesian Inference methods indicated that mitogenome sequences were useful in resolving higher-level relationship of Araneae.





Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Bernt M, Bleidorn C, Braband A, Dambach J, Donath A, Fritzsch G, Struck TH (2013) A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol 69:352–364
Blest AD, O’Carroll DC, Carter M (1990) Comparative ultrastructure of Layer I receptor mosaics in principal eyes of jumping spiders: the evolution of regular arrays of light guides. Cell Tissue Res 262:445–460
Bodner MR, Maddison WP (2012) The biogeography and age of salticid spider radiations (Araneae: Salticidae). Mol Phylogenet Evol 65:213–240
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780
Boore JL (2006) The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol 21:439–446
Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28:693–705
Cutler B (1982) Extreme northern and southern distribution records for jumping spiders (Araneae, Salticidae) in the western hemisphere. Arctic 35:426–428
Dirheimer G, Keith G, Dumas P, Westhof E (1995) Primary, secondary, and tertiary structures of tRNAs. In: Söll D, RajBhandary UL (eds) tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC, pp 93–126
Dowton M, Castro LR, Austin AD (2002) Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invertebr Syst 16:345–356
Fahrein K, Talarico G, Braband A, Podsiadlowski L (2007) The complete mitochondrial genome of Pseudocellus pearsei (Chelicerata: Ricinulei) and a comparison of mitochondrial gene rearrangements in Arachnida. BMC Genomics 8:386
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hassanin A, Léger N, Deutsch J (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa and consequences for phylogenetic inferences. Syst Biol 54:277–298
Heath TA, Hedtke SM, Hillis DM (2008) Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol 46:239–257
Hua J, Li M, Dong P, Cui Y, Xie Q, Bu W (2008) Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genomics 9:610
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kim JY, Yoon KB, Park YC (2016) The complete mitochondrial genome of the jumping spider Telamonia vlijmi (Araneae: Salticidae). Mitochondr DNA 27:635–636
Land MF (1969) Structure of the retinae of the principal eyes of jumping spiders (Salticidae: Dendryphantinae) in relation to visual optics. J Exp Biol 51:443–470
Lavrov DV, Boore JL, Brown WM (2000) The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Phylogenet Evol 17:813–824
Liu M, Zhang Z, Peng Z (2015) The mitochondrial genome of the water spider Argyroneta aquatica (Araneae: Cybaeidae). Zool Scr 44:179–190
Lopardo L, Uhl G (2014) Testing mitochondrial marker efficacy for DNA barcoding in spiders: a test case using the dwarf spider genus Oedothorax (Araneae: Linyphiidae: Erigoninae). Invertebr Syst 28:501–521
Lowe TM, Eddy SR (1997) TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Lu HF, Su TJ, Luo AR, Zhu CD, Wu CS (2013) Characterization of the complete mitochondrion genome of diurnal moth Amata emma (Butler) (Lepidoptera: Erebidae) and its phylogenetic implications. PLoS ONE 8:e72410
Lynch M (1996) Mutation accumulation in transfer RNAs: molecular evidence for Muller’s ratchet in mitochondrial genomes. Mol Biol Evol 13:209-220
Maddison WP (2015) A phylogenetic classification of jumping spiders (Araneae: Salticidae). J Arachnol 43:231–292
Maddison WP, Hedin MC (2003) Jumping spider phylogeny (Araneae: Salticidae). Invertebr Syst 17:529–549
Masta SE, Boore JL (2004) The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Phylogenet Evol 21:893–902
Masta SE, Boore JL (2008) Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol Phylogenet Evol 25:949–959
Masta SE, McCall A, Longhorn SJ (2010) Rare genomic changes and mitochondrial sequences provide independent support for congruent relationships among the sea spiders (Arthropoda, Pycnogonida). Mol Phylogenet Evol 57:59–70
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474
Palopoli MF, Minot S, Pei D, Satterly A, Endrizzi J (2014) Complete mitochondrial genomes of the human follicle mites Demodex brevis and D. folliculorum: novel gene arrangement, truncated tRNA genes, and ancient divergence between species. BMC Genomics 15:1124
Pan WJ, Fang HY, Zhang P, Pan HC (2016) The complete mitochondrial genome of pantropical jumping spider Plexippus paykulli (Araneae: Salticidae). Mitochondr DNA 27:1490–1491
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Pichaud N, Ballard JWO, Tanguay RM, Blier PU (2012) Naturally occurring mitochondrial DNA haplotypes exhibit metabolic differences: insight into functional properties of mitochondria. Evolution 66:3189–3197
Platnick NI (2016) The world spider catalog, version 15.5. American Museum of Natural History. http://research.amnh.org/iz/spiders/catalog. Accessed 10 May 2016
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Prószyński J (2016) The Salticidae (Araneae) of the World. Prepared with the assistance of the Museum and Institute of Zoology, Polish Academy of Sciences Warsaw, Poland. http://www.miiz.waw.pl/salticid/main.htm. Accessed 10 May 2016
Qiu Y, Song D, Zhou K, Sun H (2005) The mitochondrial sequences of Heptathela hangzhouensis and Ornithoctonus huwena reveal unique gene arrangements and atypical tRNAs. J Mol Evol 60:57–71
Staton JL, Daehler LL, Brown WM (1997) Mitochondrial gene arrangement of the horseshoe crab Limulus polyphemus L: conservation of major features among arthropod classes. Mol Phylogenet Evol 14:867–874
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Phylogenet Evol 30:2725–2729
Van Nieuwenhuyse P, Demaeght P, Dermauw W, Khalighi M, Stevens CV, Vanholme B, Van Leeuwen T (2012) On the mode of action of bifenazate: new evidence for a mitochondrial target site. Pestic Biochem Phys 104:88–95
Wang ZL, Li C, Fang WY, Yu XP (2014) The complete mitochondrial genome of the wolf spider Wadicosa fidelis (Araneae: Lycosidae). Mitochondr DNA 8:1–2
Wang ZL, Li C, Fang WY, Yu XP (2016) The complete mitochondrial genome of orb-weaving spider Araneus ventricosus (Araneae: Araneidae). Mitochondr DNA 27:1926–1927
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216
Xu CCY, Yen IJ, Bowman D, Turner CR (2015) Spider web DNA: a new spin on noninvasive genetics of predator and prey. PLoS ONE 10:e0142503
Yuan ML, Wei DD, Wang BJ, Dou W, Wang JJ (2010) The complete mitochondrial genome of the citrus red mite Panonychus citri (Acari: Tetranychidae): high genome rearrangement and extremely truncated tRNAs. BMC Genomics 11:597
Zhang P, Fang HY, Pan WJ, Pan HC (2016) The complete mitochondrial genome of the writing spider Argiope amoena (Araneae: Araneidae). Mitochondr DNA 27:1492–1493
Acknowledgements
This work was financially supported by Zhejiang Provincial Natural Science Foundation of China (Grant No. LQ16C140002) and National High Technology Research and Development Program of China (863 Program) (Grant No. 2012AA021601). The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wen-Yuan Fang and Zheng-Liang Wang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fang, WY., Wang, ZL., Li, C. et al. The complete mitogenome of a jumping spider Carrhotus xanthogramma (Araneae: Salticidae) and comparative analysis in four salticid mitogenomes. Genetica 144, 699–709 (2016). https://doi.org/10.1007/s10709-016-9936-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-016-9936-8