Abstract
Lipoprotein receptors, including low-density lipoprotein receptor (LDLr) relatives (Lrs) and LDLr-related proteins (Lrps), belong to the LDLr supergene family and participate in diverse physiological functions. In this study, novel sequences of lr and lrp genes expressed in the ovary of the short-finned eel, Anguilla australis, during early gonadal development are presented. The genes encoding the LDLr-like, Lrp1-like, Lrp1b-like, Lrp3, Lrp4-like, Lrp5-like, Lrp6, Lrp10, Lrp11, Lrp12-like, and Lr11-like proteins were found and identified by sequence and structure analysis, in addition to phylogenetic analysis. Genes encoding proteins previously implicated in follicle development and vitellogenin (Vtg) uptake in oviparous vertebrates were also identified, i.e. lr8 (including lr8 + and lr8- variants) and lrp13; their identification was reinforced by conserved synteny with orthologues in other teleost fish. Compared to other lr/lrp genes, the genes encoding Lr8 + , Lr8-, and Lrp13 were highly expressed in ovary during early development, decreasing as oocyte development advanced when induced by hypophysation. Furthermore, lr8 + , lr8-, and lrp13 were dominantly expressed in the ovary when compared with 17 other tissues. Finally, this study successfully detected the expression of both lr8 variants, which showed different expression patterns to those reported in other oviparous vertebrates and provided the first characterisation of Lrp13 in Anguilla sp. We propose that lr8 + , lr8-, and lrp13 encode putative Vtg receptors in anguillid eels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The low-density lipoprotein receptor (LDLr) family is represented by single-pass transmembrane proteins that share some distinctive common features with the first member discovered, the LDLr (also known as Lr7): (i) ligand-binding domains or regions with a member-specific number of LDL class A (LDLa) repeats containing the negatively charged DxSDE conserved sequence, (ii) epidermal growth factor (EGF) precursor homology domains with EGF-like repeats and YWTD repeats, and (iii) a cytoplasmic domain usually containing motifs for internalisation (e.g. FxNPxY), or for signalling (Babin et al. 2007; Schneider 2008). The family includes LDLr relatives (Lrs) and LDLr-related proteins (Lrps), of which core members in vertebrates are the LDLr and Lr8 (also known as very low-density lipoprotein receptor (VLDLr) and/or vitellogenin receptor (Vtgr)). It also includes Lrp1, Lrp1b, Lrp2, Lrp4, Lrp8, and Lrp13 (characterised in teleost fish: Reading et al. 2014), while more distant members are the Lrp3, Lrp5, Lrp6, Lrp10, Lrp11, Lrp12, and Lr11 (sorting protein-related receptor containing LDLr class A repeats (SorLA)) (Príncipe et al. 2021). Additionally, some members contain special features such as the O-linked sugar domains associated with the LDLr, Lr8, and Lrp8, or the distinct domains associated with more distant members, e.g. the vacuolar protein sorting-10 domain (VPS10) associated with Lr11, the CUB (for complement C1s/C1r, sea urchin epidermal growth factor (Uegf) and bone morphogenic protein-1 (Bmp1)) domain associated with the Lrp3, Lrp10, and Lrp12, or the MANEC domain associated with Lrp11.
In oviparous vertebrates, members of the LDLr family play important roles in oocyte development and growth, facilitating vitellogenin (Vtg) uptake. While Lr8 binds Vtg in frog (Xenopus laevis: Okabayashi et al. 1996), chicken (Gallus gallus, which also binds VLDL: Stifani et al. 1990) and various teleost fish (rainbow trout, Oncorhynchus mykiss: Davail et al. 1998; blue tilapia, Oreochromis aureus: Li et al. 2003; cutthroat trout, Oncorhynchus clarki: Mizuta et al. 2013; and white perch, Morone americana: Reading et al. 2014), recently, the novel Lrp13 member was characterised and confirmed to bind Vtg in white perch (Reading et al. 2014) and cutthroat trout (Mushirobira et al. 2015). Additionally, the lr8 gene presents two transcript variants, lr8 + and lr8-, that only differ in the presence or absence, respectively, of a region encoding an O-linked sugar domain (in frog: Okabayashi et al. 1996; chicken: Bujo et al. 1995; Senegalese sole, Solea senegalensis: Agulleiro et al. 2007; Atlantic salmon, Salmo salar: Andersen et al. 2017; blue tilapia: Li et al. 2003; cutthroat trout: Mizuta et al. 2013; rainbow trout: Prat et al. 1998; and possibly, in white perch: Hiramatsu et al. 2004; Reading et al. 2011, 2014). However, the Lr8- isoform has historically been considered the oocyte-specific Vtgr, while the Lr8 + isoform has been suggested to be a somatic receptor (Bujo et al. 1995; Mizuta et al. 2013; Prat et al. 1998).
The short-finned eel (SFE, Anguilla australis) is one of the two main temperate species of anguillid eels found in New Zealand. Anguillid eels represent a group of basal teleost fish, the Elopomorpha, which correspond to one of the earliest evolved group within the teleost lineage (Chen et al. 2015; Takezaki 2021). As such, from an evolutionary perspective, its physiology may yield insights into vitellogenin receptor biology that may be indicative of the ancestral state in teleost fish. Indeed, scarce information on Lr/Lrps is available from anguillid eels, including putative Vtgrs, to the extent that the Lrp13 member is not yet described. In addition, studies done on the SFE (Damsteegt et al. 2015a; Nguyen et al. 2020) and the European eel (A. anguilla: Jéhannet et al. 2019; Morini et al. 2020) did not detect specific transcript variants for lr8, as the qPCR primers used were not adequate to differentially amplify the lr8 + or lr8- variants. Consequently, a de novo transcriptome of SFE ovarian tissue was interrogated to examine the LDLr family members expressed in the SFE ovary during early development, with a special interest in putative Vtgrs. Two Lr8 variants, Lr8 + and Lr8-, and an Lrp13 member were found and further characterised.
Materials and methods
Sequence analysis
Novel transcript and protein sequences were retrieved from a de novo transcriptome of SFE ovarian tissue in the pre-vitellogenic stage (PV, previous to Vtg incorporation) and the early vitellogenic stage (EV, right after Vtg uptake has started). The database, presented in an earlier publication (i.e., Babio et al. 2022), was interrogated to find the Lr/Lrp members expressed in the SFE ovary. Through the stand-alone command-line application BLAST + package v2.9.0 + provided by NCBI (Camacho et al. 2009), a BLASTp search was done using Lr and Lrp amino acid sequences from other teleost fish (see species names and accession numbers in Online Resource 1). Also, SFE Lr/Lrps were searched by name in the annotated transcriptome. Then, all the associated genes that passed the filter, used to remove lowly expressed genes (Babio et al. 2022), were kept (see sequences excluded from analysis in Online Resource 1). When multiple gene IDs were annotated to the same Lr/Lrp member, only one gene ID was selected based on its highest read counts, unless it was confirmed that they corresponded to different gene sequences after nucleotide and protein alignments.
Subsequently, the corresponding protein sequences from all selected Lr/Lrp members were retrieved. The protein sequences were then subjected to BLASTp search against the nr protein database from NCBI (NCBI Resource Coordinators 2018), and gene/protein nomenclature was based on their corresponding European eel top hit (A. anguilla genome RefSeq GCF_013347855.1, Annotation Release 100, accessed in October 2021). For ease of reporting, the putative Vtgrs will be referred to as the lr8 gene encoding Lr8 +/- variants and lrp13/Lrp13, while the genes annotated to Lrp10 and SorLA will be referred to as lrp10/Lrp10 and lr11-like/Lr11-like, respectively. Protein sequences were further examined, and their conserved domains were identified using the CD-Search tool (Marchler-Bauer and Bryant 2004) from the Conserved Domain Database of NCBI (Lu et al. 2020) and the Simple Modular Architecture Research Tool (SMART: Letunic et al. 2021). Protein sequences were also subjected to phylogenetic analysis (“Phylogenetic analysis” section).
The nucleotide sequences of the putative Vtgrs from SFE were also retrieved, and their open reading frames (ORFs) were detected with the ORF finder from NCBI (available online at https://www.ncbi.nlm.nih.gov/orffinder/, accessed in August 2021). Then, their protein molecular weight was predicted using the Compute pi/MW Tool from the ExPASy server (Gasteiger et al. 2005), and signal peptides were predicted with SignalP v5.0 (Almagro Armenteros et al. 2019). Multiple protein alignments with putative orthologues from different taxa (see species names and accession numbers in Online Resources 2 and 3) were performed using Clustal Omega v1.2.4 (Madeira et al. 2019; Sievers et al. 2011).
Phylogenetic analysis
Protein sequences corresponding to Lr/Lrp members of the LDLr family from species representing mammals, amphibians, sauropsids, and teleost fish were retrieved from NCBI (see species names and accession numbers in Fig. 1) to construct a phylogenetic tree including the Lr/Lrp members found in the SFE ovary. Partial sequences (Lrp1-like, Lrp1b-like, Lrp3, Lrp10, and Lrp11) or complete protein sequences (Lr8, Lrp13, LDLr-like, Lrp4-like, Lrp6, Lrp12-like, and Lr11-like) were used (see Online Resource 1 for lengths of sequences retrieved from transcriptome). Only sequences of more than 300 amino acid residues were used; i.e. Lrp5-like was excluded from the analysis. Using MEGA v7 (Kumar et al. 2016), the sequences were first aligned with the ClustalW algorithm (Thompson et al. 1994), and the best model to describe their substitution pattern was then selected based on the lowest Bayesian information criterion (BIC). Briefly, among 56 different amino acid substitution models tested, the Jones-Taylor-Thornton (JTT) matrix-based model (Jones et al. 1992) was selected to be used with a discrete Gamma distribution (four categories, parameter G = 4.3344) to model the evolutionary rate differences among sites. Lastly, a tree was constructed with the maximum likelihood method applying 1000 bootstrap replicates. All positions with less than 95% site coverage were eliminated; thus, a total of 316 positions were kept in the final dataset. For visualisation purposes, the Lrp11 group was used as a relative outgroup, similar to Mushirobira et al. (2015).
Genomic synteny of putative vitellogenin receptors
Due to the lack of SFE genomic data, the surrounding genomic arrangements of the predicted lr8 and lrp13 genes for European eel were examined and compared with other teleost fish, using the genomic context section from Entrez Gene (NCBI’s database for gene-specific information, Maglott et al. 2011). The sequences used from the European eel corresponded to the genes encoding the BLASTp top hit using SFE putative Vtgrs as query sequences, i.e. A. anguilla genome RefSeq GCF_013347855.1, Annotation Release 100 (accessed in October 2021). The predicted lr8 orthologues were compared between the European eel (gene ID 118,212,928), zebrafish (gene ID 393,897), and Nile tilapia (gene ID 100,703,410), while predicted lrp13 orthologues were compared between the European eel (gene ID 118,237,195), zebrafish (gene ID 562,438), and medaka (Orzyas latipes, gene ID 101,159,109).
Tissue distribution of putative vitellogenin receptors
The transcript abundances of the lr8 variants (two lr8 transcript variants were found, see “Primer design” and “Sequence analysis and phylogenetics” sections) and lrp13 were estimated in 18 tissues (i.e. red muscle, white muscle, posterior kidney, gill, ovary, anterior kidney, spleen, anterior gut, posterior gut, thyroid, heart, liver, eye, pituitary, head kidney, forebrain, hindbrain, and midbrain) using qPCR (“PCR and qPCR reactions” section) after RNA extraction and cDNA synthesis (“RNA extraction, DNase treatment, and cDNA synthesis” section). All tissues were collected from three wild-caught female eels in the EV stage (Lake Ellesmere, New Zealand; capture year 2020; gonadosomatic index – GSI = 2.54 ± 0.1%, as described previously in Falahati et al. (2021)).
Expression of putative vitellogenin receptors during ovarian artificial maturation
The ovarian artificial maturation experiment was carried out by Damsteegt et al. (unpublished data) and Mercuriali et al. (unpublished data). Briefly, thirty-five EV stage eels (capture year 2017) were split between a 1000-L recirculating treatment tank (n = 30) and a 200-L recirculating control tank (n = 5) both maintained at 30–35 ppt, 16–17 °C, 12/12 light/dark regime. Treatment eels received weekly intramuscular injections of salmon pituitary homogenate (SPH, 10 mg/kg, as in Damsteegt et al. 2020), while control eels received weekly injections of eel Ringer’s solution. Before starting the injections, a group of non-treated eels (time point zero (week 0), n = 5) was euthanised with an overdose of benzocaine (0.3 g/L, Sigma-Aldrich) and dissected. Similarly, SPH-treated eels were euthanised every 2 weeks, for a maximum of 10 weeks, to detect the progression of oogenesis (n = 4–5 per sampled week). Finally, the Ringer-treated eels were euthanised after 10 weeks of treatment (control group – C –, n = 5). During a routine water change at week 7, the water quality of the tanks was affected, resulting in deaths and reduced sample sizes for week 8 and week 10 (n = 4).
During dissections, total body weight and ovary weight were measured to calculate the GSI. Fragments of ovarian tissue were either flash-frozen for RNA extractions (“RNA extraction, DNase treatment, and cDNA synthesis” section) and downstream analysis (qPCR, “PCR and qPCR reactions” section), fixed in Bouin’s for histological analysis, or kept in eel Ringer’s solution for measurement of oocyte diameter (OD) later that same day. Fixed ovarian fragments were dehydrated and embedded in paraffin, sectioned at 5 μm using a Leica RM2125RT microtome (Leica Biosystems, Nussloch, Germany), and stained with hematoxylin and eosin (Damsteegt et al. 2014; Lokman et al. 2016). Micrographs were captured with an Olympus camera SC100 and an Olympus adaptor U-TVO.5XC-3 attached to an Olympus microscope BX51 (Olympus Corporation, Tokyo, Japan). The OD was measured on ovarian fragments subjected to collagenase digestion (1 mg/ml, Lokman et al. 2015). Once isolated follicles were obtained, photographs were taken using an Olympus stereo microscope SZX2-ILLD attached to an Olympus camera SC100 and an Olympus adaptor U-TVO.5XC-3, and images were analysed using the imaging software Olympus CellSens Standard (Olympus Corporation, Tokyo, Japan). In keeping with Lokman et al. (2007), data from the 50% highest-ranked oocytes were retained to calculate the average OD.
RNA extraction, DNase treatment, and cDNA synthesis
Frozen samples from the tissue distribution assay and the artificial maturation experiment were subjected to total RNA extraction using TRIzol™ Reagent (Invitrogen, Thermo Fisher Scientific Inc.) following the manufacturer’s instructions. Then, 5 μg of total RNA from each sample was treated with DNase TURBO DNA-free™ kit (Invitrogen, Thermo Fisher Scientific Inc.), of which 500 ng was reverse-transcribed with PrimeScript™ RT reagent kit (Takara Bio Inc., Shiga, Japan) using Oligo(dT) and random hexamer primers. Samples were diluted with Milli-Q water (MQW) to 10 ng/μl to be used in qPCR assays (“PCR and qPCR reactions” section).
The cDNA amplified by PCR for subsequent tenfold serial dilution, to construct qPCR standard curves (“Primer design” section), was obtained from an ovarian fragment of one EV eel (capture year 2019: Babio et al. 2022). This sample was subjected to total RNA extraction using the NucleoSpin RNA kit (Macherey–Nagel, Düren, Germany) following the manufacturer’s instructions. The sample was then reverse-transcribed with SuperScript™ IV (Invitrogen, Thermo Fisher Scientific Inc.) using Oligo(dT) primers and used in PCR assays (“PCR and qPCR reactions” section).
Primer design
The PCR primers employed to generate templates to use as qPCR standards and the qPCR primers used to quantify the β-actin (actb) and elongation factor-1α (eef1a) transcripts were validated in previous studies (Table 1). PCR and qPCR primers used to amplify lr8 variants and lrp13 were designed with Primer3web software v. 0.4.0 (Koressaar and Remm 2007; Untergasser et al. 2012), using the corresponding retrieved sequences (“Sequence analysis” section). To generate qPCR standard curves, the complete ORF from lr8 variants and lrp13 were amplified using specific PCR primers (Table 1). Two lr8 splice variants were found in the SFE, which were amplified using a common pair of primers and then separated based on size selection after electrophoresis. All PCR amplicons were electrophoresed to verify their correct size and gel-extracted using the NucleoSpin gel and PCR clean-up kit (Macherey–Nagel, Düren, Germany) following the manufacturer’s instructions. Once the identity of the PCR products was verified (Sanger sequencing, Genetic Analysis Services, University of Otago), they were subjected to ten-fold serial dilutions in MQW to construct qPCR standards.
Specific qPCR primer pairs were designed to amplify products between 100 and 200 bp for both lr8 variants and lrp13 (Table 1). To detect the expression of the lr8 + variant, qPCR primers were designed to target the region encoding the putative O-linked sugar domain. In contrast, to detect the lr8- variant, the reverse primer was designed to target the site of this missing domain, i.e. spanning the corresponding exon boundary obtained after splicing (see Online Resource 4). All qPCR amplicons were electrophoresed, gel-extracted, and sequenced to corroborate their identity, as described above.
PCR and qPCR reactions
All PCR reactions were carried out on an Eppendorf Mastercycler PCR machine. To amplify ORFs encoded by lr8 variants and lrp13, the MyFi Mix (Bioline, Meridina Bioscience, London, UK) was used. Initial denaturation at 95 °C for 2 min was followed by 40 cycles of denaturation at 95 °C for 30 s, annealing for 30 s (see Table 1 for primer-specific annealing temperatures), and extension at 72 °C for 2 min (lr8 variants) or 2.7 min (lrp13). Afterwards, a final extension at 72 °C for 5 min was done. The actb and eef1a PCR amplifications were done using the MangoTaq™ DNA polymerase kit (Bioline, Meridina Bioscience, London, UK). In this case, the cycling conditions involved an initial denaturation step at 95 °C for 5 min, and 35 cycles of 95 °C for 30 s, annealing for 30 s (see Table 1 for primer-specific annealing temperatures), and extension at 72 °C for 1 min. Subsequently, a final extension at 72 °C for 5 min was done.
All qPCR reactions were carried out on a QuantStudio™ 5 (Applied Biosystems, Thermo Fisher Scientific Inc.), and the results were analysed using QuantStudio™ Design and Analysis Software (Applied Biosystems, Thermo Fisher Scientific Inc.). The reactions were prepared with SYBR® Premix Ex Taq™ II (Takara Bio, Kyoto, Japan) following the manufacturer’s instructions. The cycling conditions started with a hold step at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing for 10 s (see Table 1 for primer-specific annealing temperatures), and extension at 72 °C for 5 s. Finally, a melt curve analysis was carried out (95 °C for 1 s, 60 °C for 20 s, and 95 °C for 1 s). Samples were run in duplicate, and where possible all samples, no-template controls and standards were run on the same 96-well qPCR plate. Due to insufficient space, samples from the tissue distribution assay were divided between two plates and run with inter-assay quality controls (two sets of samples amplified on both plates) to determine the coefficient of variation (CV) between runs. Assay efficiencies for the tissue distribution assay were between 97.9 and 102.2% with a CV of 6.4% (lr8-), between 92.2 and 93.7% with a CV of 16.0% (lr8 +), and between 95.1 and 95.2% with a CV of 12.5% (lrp13). Assay efficiencies for the artificial maturation experiment were 95.0% (lr8-), 95.2% (lr8 +), 96.7% (lrp13), 95.3% (actb), and 95.1% (eef1a).
Normalisation of qPCR data
The tissue distribution data were normalised over total RNA. Likewise, after unsuccessful attempts to find suitable reference genes to normalise the expression of target genes during the progression of oocyte development (ovarian artificial maturation experiment), the data were normalised over total RNA. The genes actb and eef1a were tested and unstable expression across the different developmental stages prevented their use as reference genes (Online Resource 5). In addition, the qPCR data from the ovarian artificial maturation experiment were also analysed with the Markov chain Monte Carlo approach (Matz et al. 2013), showing similar results when compared to the data normalised over total RNA (data not shown). Therefore, the data shown are the normalised over total RNA. The total RNA concentration following DNase treatment was measured using Qubit™ RNA broad range assay kit (Invitrogen, Thermo Fisher Scientific Inc.).
Statistical analysis
Data are presented as mean ± SEM. Statistical analyses were only performed on the qPCR data obtained from the artificial maturation experiment. Data were tested for normality and homoscedasticity using the Shapiro–Wilk test modified by Rahman and Govindarajulu (1997) and the Levene’s test (Levene 1960), respectively. When assumptions were violated, data were either log-transformed (lr8- and actb transcript abundances normalised over total RNA, and lr8- transcript abundance normalised over actb) or analysed with non-parametric tests (eef1a transcript abundance normalised over total RNA). Except for this last dataset, each variable was tested using a one-way ANOVA, followed by the Scheffé post-hoc test (Scheffé 1953) due to unequal sample sizes, comparing weeks 0, 2, 4, 6, 8, and 10. Additionally, week 0 and control group C were compared with independent t-tests using an adjusted p value using the Bonferroni correction. Kruskal Wallis, comparing weeks 0, 2, 4, 6, 8, and 10, or Mann–Whitney U test, comparing week 0 and control group C, were used to analyse eef1a transcript abundance normalised over total RNA. All analyses were done using InfoStat v.2018 (Di Rienzo et al. 2018).
Results
Sequence analysis and phylogenetics
Based on the searches of the transcriptome database (Online Resource 1), the genes encoding the proteins Lr8, Lrp13, LDLr-like, Lrp1-like, Lrp1b-like, Lrp3, Lrp4-like, Lrp5-like, Lrp6, Lrp10, Lrp11, Lrp12-like, and Lr11-like were expressed in the ovary of the SFE during early development (PV and EV stages) (Table 2). The analysis of conserved domains from the deduced protein sequences, along with their alignment with corresponding LDLr family members from different taxa, supports their identity and classification (data only shown for Lr8 and Lrp13 in Online Resources 2 and 3, respectively). Additionally, these members grouped into distinct clusters with their counterparts from different taxa when a phylogenetic tree was constructed, except for Lrp6 which grouped with both Lrp6 and Lrp5 members (Fig. 1).
Phylogenetic tree of Lr/Lrp members of the LDLr family from different taxa (accession numbers and species names are shown), including the members expressed in the ovary of the short-finned eel, Anguilla australis, denoted by their TRINITY identification in bold red. The tree was constructed using the maximum likelihood method based on the Jones-Taylor-Thornton (JTT) matrix-based mode with a discrete Gamma distribution (G = 4.3344), applying 1000 bootstrap replicates. Bootstrapping values (%) are shown at tree nodes
Two genes encoding putative Vtgrs (Lr8 and Lrp13) were found, and their nucleotide and protein sequences were further examined. Based on sequence similarity to other vertebrate Vtgrs, the gene TRINITY_DN701_c1_g1 was designated as lr8; two splice variants (TRINITY_DN701_c1_g1_i4 and TRINITY_DN701_c1_g1_i9) of this gene were detected and designated as Lr8 + and Lr8- isoforms, depending on the presence or absence, respectively, of a putative O-linked sugar domain (Online Resource 2). The lr8 + and lr8- variants display a 5′-untranslated region (UTR) of 78 bp; ORFs of 2700 bp and 2595 bp, respectively; and a 3′-UTR of 399 bp. The corresponding Lr8 + and Lr8- deduced protein sequences have a length of 899 and 864 amino acids with predicted molecular weights of 99.04 kDa and 95.36 kDa, respectively. Similarly, even though it was functionally annotated as an Lrp4 member (Table 2), the gene TRINITY_DN2157_c0_g1 was designated as lrp13 due to its structural domains, i.e. seven LDLa repeats at the N-terminal and a unique C-terminal domain containing an extra LDLa repeat (Online Resource 3). The lrp13 sequence contains a 5′-UTR of 51 bp, an ORF of 3705 bp, and a 3′-UTR of 83 bp, encoding a protein of 1234 amino acids with a predicted molecular weight of 132.76 kDa.
The most likely orthologues of the SFE lr8 and lrp13 genes found among the predicted sequences of the European eel (Table 2) were used to examine and compare their syntenic arrangements with other teleost species (see “Genomic synteny of putative vitellogenin receptors” section). The proteins encoded by both genes in both species (SFE lr8 versus European eel vldlr (gene ID 118,212,928) and SFE lrp13 versus European eel LOC118237195) share a high percentage of identity, supporting their inferred orthology. While SFE Lr8 + and European eel VLDLr isoform X14 share 99.4%, SFE Lr8- and European eel VLDLr isoform X19 share 99.5%. SFE Lrp13 and European eel proLrp1-like share 95.9%.
Genomic synteny of putative vitellogenin receptors
Both the lr8 and lrp13 genes share genomic synteny when compared with corresponding orthologues from different teleost fish (Fig. 2). Using the NCBI database, the predicted lr8 (vldlr) and lrp13 (LOC118237195) genomic arrangements for European eel were compared between zebrafish and Nile tilapia and zebrafish and medaka, respectively. For both genes, similar arrangements have been found in the species compared, showing with few exceptions, a retention of direction of the reading frames and the same adjacent genes. Most importantly, the SFE lr8 and lrp13 counterparts in the European eel showed similar conserved syntenic arrangements as their orthologues in other teleost fish. In all species compared, the lr8 gene is consistently surrounded by the genes secreted phosphoprotein 1 (spp1) and SH3-domain-binding protein 2 (sh3bp2) (upstream) and potassium channel subfamily V member 2a (kcnv2a) and pumilio RNA-binding family member 3 (pum3) (downstream). The adjacent genes downstream of lrp13 are slightly more variable between the species compared, but the upstream gene ring finger and CHY zinc finger domain containing 1 (rchy1) is present in each species.
Genomic context of lr8 and lrp13 genes in different teleost fish based on the information derived from NCBI database. The European eel (A. anguilla), zebrafish (D. rerio), medaka (O. latipes), and Nile tilapia (O. niloticus) were compared. The chromosome number and the regions analysed are indicated for each species. Genes are represented by arrows and named when appropriate. lr8 and lrp13 genes are represented by red arrows. spp1, secreted phosphoprotein 1; sh3bp2, SH3-domain-binding protein 2; vldlr/lr8; kcnv2a, potassium channel subfamily V member 2a; pum3, pumilio RNA-binding family member 3; rchy1, ring finger and CHY zinc finger domain containing 1; lrp13/LOC118237195/vldlr-like/proLrp1-like; smad2, SMAD family member 2; susd1, sushi domain containing 1; kcnv2b, potassium channel subfamily V member 2b
Tissue distribution of putative vitellogenin receptors
All putative Vtgrs, i.e. lr8 + , lr8-, and lrp13, were highly expressed in the SFE ovary during the EV stage when compared with 17 somatic tissues (Fig. 3). The three transcripts also showed a low expression in white muscle.
Expression of putative vitellogenin receptor genes during artificial maturation
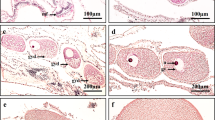
The oocyte development during artificial maturation was examined through ovarian histological sections (Fig. 4). During SPH treatment, oocytes started to actively accumulate great amounts of yolk proteins (Vtg accrual), which directed the progress from the EV stage to the late vitellogenic (LV) stage (GSI and OD for week 2 = 4.8 ± 0.5% and 268.2 ± 10.8 μm; week 4 = 6.2 ± 0.8% and 341.2 ± 29.3 μm; week 6 = 11.9 ± 1.5% and 446.7 ± 16.6 μm; week 8 = 6.4 ± 1.3% and 386.9 ± 16.5 μm; week 10 = 23.2 ± 6.1% and 695.0 ± 75.5 μm). In contrast, ovaries from the control groups (week 0 – GSI = 2.2 ± 0.1% and OD = 221.5 ± 4.3 μm – and C – GSI = 2.0 ± 0.1% and OD = 214.9 ± 6.7 μm) were representative of early development, containing both oocytes in the PV and either the late PV stage or EV stage, although no evident yolk accumulation was noted.
Micrographs of ovaries from short-finned eel, Anguilla australis, during artificial maturation stimulated by weekly injections of salmon pituitary homogenate (10 mg/kg) at b week 2, c week 4, d week 6, e week 8, or f week 10. a Non-treated eels at week 0 and g eels injected weekly with Ringer’s solution for 10 weeks were used as control groups. The 100 μm scale bars are shown. * indicate oil droplets, arrows indicate yolk proteins, and PV denotes pre-vitellogenic oocytes
The relative expression of lr8 + , lr8-, and lrp13 normalised over total RNA decreased throughout progression of oogenesis induced by SPH treatment (one-way ANOVA: lr8 + : F = 68.06, df = 5, p < 0.0001; lr8-: F = 15.31, df = 5, p < 0.0001; lrp13: F = 24.04, df = 5, p < 0.0001) (Fig. 5). While no significant differences were found between control groups, i.e. week 0 and C groups, for lr8- and lrp13, lr8 + transcript abundance in group C was significantly lower than that at week 0 (t-test: lr8 + ; t = 3.43, df = 8, p < 0.01). On the contrary, if the data are normalised over actb, the relative expression of lr8 + , lr8-, and lrp13 remains stable during artificial maturation, showing no significant differences in transcript abundance between the experimental groups (p > 0.05 for both one-way ANOVA and t-test) (see Online Resource 5).
Relative transcript abundance of a lr8 + , b lr8-, and c lrp13 during induced maturation in the ovary of short-finned eel, Anguilla australis. Data are shown as mean ± SEM for females terminally sampled at fortnightly intervals and for Ringer’s-injected control – C – (all groups at n = 5, except for weeks 8 and 10 at n = 4). Different letters indicate significant differences between groups (p < 0.05; one-way ANOVA: weeks 0, 2, 4, 6, 8, 10). No significant differences were found between week 0 and C groups, except for lr8 + (t-test, * indicates significant difference)
Discussion
Lr/Lrp members expressed in ovary during early development
Based on the identification of conserved domains, multiple sequence alignments, and the phylogenetic analysis, several Lr/Lrp members of the LDLr family expressed in the SFE ovary during early development were identified. In addition to the genes encoding putative Vtgrs (Lr8 and Lrp13), the genes encoding Ldlr-like, Lrp1-like, Lrp1b-like, Lrp3, Lrp4-like, Lrp5-like, Lrp6, Lrp10, Lrp11, Lrp12-like, and Lr11-like were found. Since disparate levels of gene expression were detected among them, in comparison to the highly expressed putative Vtgrs (based on the read counts obtained from the transcriptome database, see Table 2), additional research will be needed to elucidate the biological relevance of their expression in the ovary of anguillid eels during early ovarian development, and the functions they may play in teleost fish in general. After phylogenetic analysis, the SFE Lrp6 grouped with both Lrp5 and Lrp6 members from different taxa, likely due to their high homology (Ren et al. 2021). Even though complete Lr/Lrp protein sequences were found for some (i.e. Lr8, Lrp13, LDLr-like, Lrp4-like, Lrp6, Lrp12-like, and Lr11-like), others presented with partial protein sequences (i.e. Lrp1-like, Lrp1b-like, Lrp3, Lrp 5-like, Lrp10, and Lrp11). This possibly affected the phylogenetic analysis, since after alignment, all positions with less than 95% site coverage were eliminated, and a total of 316 residues were considered for the analysis. As putative Lrp5-like, containing only 146 amino acid residues, was excluded from the phylogenetic analysis, it is essential to obtain its whole sequence so as to align it with other Lr/Lrps and corroborate its identification. Additional research is warranted to clarify this.
Receptors from the LDLr family play diverse functions in lipid metabolism, cell homeostasis, and signal transduction pathways (Dieckman et al. 2010; Li et al. 2001; Schneider and Nimpf 2003), being involved in receptor-mediated endocytosis of various ligands, e.g. lipoproteins, hormones, protease inhibitor complexes, vitamins, extracellular matrix proteins, growth factors, and signalling molecules (Dieckman et al. 2010; May et al. 2007). To date, little is known about the function that many of these receptors may play in teleost fish, especially during ovarian development. Indeed, most of the available data on Lr/Lrp function comes from mammals, with a special focus on development and human pathologies. For instance, Lrp1 is widely expressed in several tissues and can play a variety of physiological functions from lipoprotein metabolism, clearance, and degradation of proteases to signalling pathways and development (Lillis et al. 2008). Lrp1b has been mainly implicated in roles in the cell cycle, cellular growth regulation, and proliferation in the context of cancer (Dieckman et al. 2010), similarly to Lrp11, which has been related to oncogenesis and the stress response (Gan et al. 2020; Wang et al. 2019; Xu et al. 2014). Little is known about the CUB domain-containing members Lrp3, Lrp10, and Lrp12, but they have been primarily related to central nervous system development (Cuchillo-Ibañez et al. 2021; Pohlkamp et al. 2017). Also, the SorLA/Lr11 member has been implicated in Alzheimer’s disease pathogenesis (Dieckman et al. 2010). The widely co-expressed paralogous genes encoding Lrp5 and Lrp6 are components of the Wnt signalling pathway, functioning as co-receptors of Wnt ligands (Ren et al. 2021). In addition, Lrp4 has a role in cellular signal transduction regulating the Wnt signalling pathway and also in interacting with other signalling molecules, affecting bone morphogenesis, tooth development, and neuromuscular junction development (see Dieckman et al. 2010 and references therein). In mammals, the Wnt signalling pathway is known to be involved in embryonic development and ovarian differentiation and development, among other functions (Harwood et al. 2008; Tevosian and Manuylov 2008; Zheng et al. 2006). Correspondingly, in rainbow trout, Nicol and Guiguen (2011) detected the expression of several Wnt pathway genes not only during gonadal differentiation, but also during gametogenesis, suggesting the possible implication of this signalling pathway in teleost fish folliculogenesis and/or oogenesis.
Due to their shared ligand-binding properties, many members of the LDLr family can participate in the endocytic uptake of lipoproteins, playing important roles in lipoprotein metabolism and lipid homeostasis. The recognition of circulating lipoproteins is possible through interaction with apolipoproteins (Apos), the protein components of lipoprotein complexes determining also their assembly, structure, and transport (Jonas and Phillips 2008). While the mammalian LDLr, and likely Lrp6 as well (Go and Mani 2012; Ye et al. 2012), bind both ApoE and ApoB (Esser et al. 1988; Gui et al. 2016), the Lr8, Lrp1, and Lr11 members only bind ApoE (Gui et al. 2016; Taira et al. 2001; Zhao et al. 2018). Whereas ApoE and Vtg have long been suggested to be functional analogues (Steyrer et al. 1990), ApoB and Vtg share structural similarities as they both belong to the large lipid transfer protein (LLTP) superfamily (Babin et al. 1999). Notably, the chicken Lr8 is capable of binding both Vtg and VLDL, an ApoB-containing lipoprotein (Stifani et al. 1990), as well as the mammalian ApoE (Steyrer et al. 1990), and blue tilapia’s Vtg has a similar receptor-binding region to ApoB and ApoE (Li et al. 2003). Consequently, it is possible that various Lr/Lrps may be involved in egg yolk formation during oocyte development due to structural similarities between ligands and receptors, respectively.
In teleost species laying fatty eggs, egg yolk formation relies on the uptake and accumulation of Vtg and neutral lipids harboured within low-density lipoproteins, e.g. supplementation of incubation media with VLDL results in lipid uptake in vitro by ovarian fragments of anguillid eels (Endo et al. 2011; Damsteegt et al. 2015b). To date, it is known that whereas the Vtgrs are implicated in egg yolk formation by assisting in Vtg uptake, LDLr likely contributes to neutral lipid incorporation during early development. Although the exact mechanism is unclear, Damsteegt et al. (2015b) showed that the predicted LDLr may be a major player in fatty acid accumulation in A. australis. Of note, the human Lrp6 was shown to also regulate LDLr-mediated LDL uptake in vitro, besides its known function as a co-receptor of Wnt ligands (Go and Mani 2012; Ye et al. 2012). In accordance with this, other Lr/Lrps could conceivably assist the main receptors in lipoprotein binding and/or uptake. Nevertheless, it is still unknown if other LDLr family members with similar binding properties, like Lrp1, Lr11, and Lrp6, are implicated in egg yolk formation in teleost fish. Indeed, future research on the Lr/Lrps expressed in the ovary of teleost fish is warranted to elucidate their functions during oocyte development.
Notably, Damsteegt et al. (2015a) analysed the expression of the predicted SFE LDLr during early oogenesis. However, the current ovarian transcriptome database did not yield evidence for ldlr expression and instead only showed the expression of a gene encoding a receptor protein closely related to LDLr, the TRINITY_ DN13732_c0_g1 gene encoding the LDLr-like protein. This member was grouped within the Lr7 cluster due to sequence similarity, although it does not seem to correspond to an orthologue of the LDLr; i.e. it only shares 73.75% identity with the predicted European eel LDLr (accession number XP_035262743.1/associated to the ldlrb gene), which is orthologous to the zebrafish and human LDLrs. Instead, it was linked to the LDLr-like protein in European eel (accession number XP_035254684.1/associated to the LOC118217039 gene), sharing 96.25% identity. In addition, the gene TRINITY_DN113818_c0_g1, also annotated as LDLr, was found to share 100% identity with the predicted SFE LDLr partial sequence (Damsteegt et al. 2015a) and 98.68% identity with the predicted European eel LDLr. Yet, this gene was excluded from the analysis as it was filtered out due to a low number of reads. After alignment (data not shown), and using the European eel genomic data from NCBI as a reference, both SFE LDLr-like and LDLr seem to be products of different genes, possibly paralogues. This implies that more than one member from the Lr7 group, i.e. LDLr family member containing seven LDLa repeats in its ligand-binding domain, are expressed in the SFE ovary during early development. Targeted studies should be aimed at elucidating the specifics of each gene/protein and their contribution to oocyte development.
Characterisation of putative vitellogenin receptors
Two genes encoding LDLr family members involved in Vtg binding in some teleost fish, known as lr8 and lrp13, were confirmed to be expressed in the SFE. Lr8 + and Lr8- protein isoforms generated by alternative splicing of transcripts derived from the lr8 gene were also identified. Both isoforms contain eight LDLa repeats in the ligand-binding domain and only differ in the presence or absence of a putative O-linked sugar domain (Lr8 + and Lr8-, respectively). Similar to rainbow trout, both transcript variants differ in 105 bp encoding 35 amino acids representing a putative O-linked sugar domain, which presents low levels of homology and glycosylation when compared to the chicken Lr8 + and the human Lr8 + (Prat et al. 1998). According to the predicted European eel lr8 gene sequence (vldlr, gene ID 118,212,928), this region would be encoded by exons 25 and 26, differing from chicken and human O-linked sugar domain-coding regions that only span one exon (Bujo et al. 1995; Magrané et al. 1999). Similarly, the rainbow trout O-linked sugar domain appears to be encoded by two exons, implying that it may be a common feature in some teleost fish (cf. rainbow trout predicted lr8 gene sequence NCBI Gene ID: 100,136,065 and regions covered by Lr8 + and Lr8- isoforms—accession numbers XP_036792567.1 and XP_021478075.2, respectively).
Lrp13 proteins found in teleost fish have a species-specific number of N-terminal ligand-binding repeats (7–13) and a unique C-terminal LDLa repeat (+ 1) (Hiramatsu et al. 2013, 2015). For instance, Lrp13 proteins were found in striped bass (Morone saxatilis) and white perch (7 + 1) (Reading et al. 2014), cutthroat trout (13 + 1) (Mushirobira et al. 2015), greater amberjack (Seriola dumerili) (10 + 1) (Pousis et al. 2019), and yellow croaker (Larimichthys crocea) (7 + 1) (Gao et al. 2020). The Lrp13 member expressed in the SFE presents seven LDLa repeats at the N-terminal region and the typical C-terminal configuration of other known Lrp13 proteins. Additional support for the identification of the putative Vtgrs found in the SFE was obtained when examining the syntenic arrangements of the corresponding coding genes using the European eel reference genome. Both the lr8 and lrp13 genes share the same syntenic arrangements as their corresponding predicted orthologues from different teleost fish, similar to what was found by Reading et al. (2014) when comparing the zebrafish and Nile tilapia lr8 and lrp13 genes, by Andersen et al. (2017) when comparing lr8 genes from Atlantic salmon, three-spined stickleback, African coelacanth (Latimeria chalumnae), spotted gar (Lepisosteus oculatus), and elephant fish (Callorhinchus milii), and by Wang et al. (2017) when comparing lrp13 genes from Chinese tongue sole, medaka, yellow croaker, and Japanese puffer.
After identification and molecular characterisation, the putative Vtgrs from the SFE were further studied, examining their transcript abundance in different somatic tissues. All three putative Vtgrs were expressed almost exclusively in the ovary when compared to 17 somatic tissues from three EV eels, although presenting low expression in white muscle as well. Low expression of lr8 and lrp13 genes in muscle has been previously detected in other teleost fish (lr8: Gao et al. 2020; Li et al. 2003; Mizuta et al. 2013; Prat et al. 1998 and lrp13: Gao et al. 2020; Mushirobira et al. 2015; Reading et al. 2014). Although it remains unclear what role they may play in this tissue and whether their level of expression is significant for physiological function, these receptors may play a part in lipid metabolism facilitating the energy demands of the tissue.
In teleost fish examined to date, the expression of lrp13 is predominant in ovary (in yellow croaker: Gao et al. 2020; cutthroat trout: Mushirobira et al. 2015; striped bass: Reading et al. 2014; Chinese tongue sole and medaka: Wang et al. 2017). While lr8 also shows high expression in ovary, its +/- variants usually show differential expression; i.e. the lr8- variant is dominant in the ovary in comparison to the lr8 + variant (e.g. in Senegalese sole: Agulleiro et al. 2007; Atlantic salmon: Andersen et al. 2017; blue tilapia: Li et al. 2003; cutthroat trout: Mizuta et al. 2013; rainbow trout: Prat et al. 1998). Surprisingly, in the ovary of the SFE, the expression of the lr8 + variant was higher than the expression of the lr8- variant. In every qPCR assay performed (to test primers using cDNA pool from three EV eels, capture year 2019; tissue distribution on three EV eels, capture year 2020; artificial maturation experiment with EV eels, capture year 2017), the lr8 + variant was consistently detected between 1 and 2 Ct values lower than the lr8- variant (data not shown). Notably, higher expression could imply higher protein abundance, and thus, protein abundance data should clarify the significant contribution of each receptor to the final Vtgr pool. Whether this expression pattern found in A. australis, a basal teleost, reflects the ancestral state and the predominant expression of lr8- in ovary from other teleost fish represents the derived state requires further investigation. Additionally, since the only difference between isoforms is the presence of a putative O-linked sugar domain in Lr8 + , which in mammalian receptors are thought to confer cell surface stability protecting against shedding proteases (in Lr8 + : Iijima et al. 1998; Magrané et al. 1999; LDLr: Kozarsky et al. 1988; in Lrp8 + : May et al. 2003; Wasser et al. 2014), it may differentially affect the availability of the receptors in the oocyte membrane during vitellogenic growth. Although there are no data available from teleost fish to support this suggestion, it deserves further examination, especially when considering the difference in the levels of glycosylation between teleost fish and mammalian homologues which may affect the function of the domain.
Lastly, lr8 + , lr8-, and lrp13 gene expression showed the same decreasing trend during ovarian artificial maturation. The expression of the control group in which fish were injected weekly with eel Ringer’s solution for 10 weeks was likely affected by captivity, as it showed lower and/or similar values than week 0 and week 2, although only significant differences were found in lr8 + transcript abundance. The decreasing expression pattern of Vtgrs during oocyte development has been reported in other teleost fish (e.g. in Atlantic salmon lr8: Andersen et al. 2017; largemouth bass, Micropterus salmoides lr8-: Dominguez et al. 2012; yellow croaker lr8- and lrp13: Gao et al. 2020; cutthroat trout lr8-: Mizuta et al. 2013; striped bass lr8- and lrp13: Reading et al. 2014). However, it is most likely that the changes in RNA composition during oocyte development affect the relative expression of target genes. Indeed, the expression of target genes could be masked or diluted predominantly as a result of a decrease in the mRNA/total RNA ratio during the later stages of development, in part, due to higher expression of 18S and 28S rRNA (Kroupova et al. 2011). Also, the increase by orders of magnitude of specific transcripts during development, such as mitochondrial cytochrome b (Lokman et al. 2003), and conceivably, all other protein-encoding mitochondrial genes, and the accumulation of maternal transcripts during the early stages of oocyte development (Lubzens et al. 2017) could contribute to this effect. Similarly, the dramatic increase in oocyte size during development reflects an increase in cytoplasmic volume promoting the dilution effect. Taken together, this could explain the decreasing trend of expression of the genes analysed, including that of the normaliser genes actb and eef1a, when oocyte development advances. When data were standardised over total RNA, the relative expression of target genes decreased as the developmental stage advanced, whereas when using actb, their expression remained stable. In any case, previous data from other teleost fish affirm that these expression profiles are commonly seen in other teleost fish as Vtgrs either decrease when development advances (see references above) or remain stable (Morini et al. 2020). Physiologically, both scenarios are reasonable as the Vtgrs are likely recycled back to the membrane surface after endocytosis, without de novo synthesis of Vtgr transcripts during vitellogenic growth.
Conclusions
Multiple Lr/Lrp members of the LDLr family, including putative Vtgrs, are expressed in the SFE ovary during early development, suggesting they play roles in this tissue. The functional redundancy of Lr/Lrp members (Schneider et al. 1999) may indicate that (some of) these roles can overlap between multiple members, likely contributing to key steps in oocyte development and possibly assisting in egg yolk development. However, the possible functions that the Lr/Lrp members play in supporting egg yolk formation and/or cellular signalling pathways involved in oocyte development need to be further explored in the SFE and teleost fish in general. Additionally, it has to be determined if their level of expression has any biological significance. Finally, three putative Vtgr receptors, i.e. Lr8 + , Lr8-, and Lrp13, known to be involved in Vtg uptake and egg yolk formation in some fish, which ultimately translates into egg quality and reproductive effort, have been characterised in the SFE. The three genes are mainly expressed in ovary when compared to somatic tissues, and their expression decreases as oocyte development advances by artificial means. The expression pattern found in the SFE, in which both lr8 variants are mainly expressed in ovary and lr8 + expression is higher than lr8-, is in contrast to what was detected in other oviparous vertebrates. Further studies in anguillid eels are needed to confirm this pattern and specify the contribution of each variant to the final Vtgr pool available for Vtg uptake.
Data Availability
The data presented in this study is contained within the article and supplementary material.
Change history
23 February 2023
Missing Open Access funding information has been added in the Funding Note.
References
Agulleiro MJ, André M, Morais S, Cerda J, Babin PJ (2007) High transcript level of fatty acid-binding protein 11 but not of very low-density lipoprotein receptor is correlated to ovarian follicle atresia in a teleost fish (Solea senegalensis). Biol Reprod 77:504–516. https://doi.org/10.1095/biolreprod.107.061598
Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. https://doi.org/10.1038/s41587-019-0036-z
Andersen Ø, Xu C, Timmerhaus G, Kirste KH, Næve I, Mommens M, Tveiten H (2017) Resolving the complexity of vitellogenins and their receptors in the tetraploid Atlantic salmon (Salmo salar): ancient origin of the phosvitin-less VtgC in chondrichthyean fishes. Mol Reprod Dev 84:1191–1202. https://doi.org/10.1002/mrd.22881
Babin PJ, Bogerd J, Kooiman FP, Van Marrewijk WJ, Van der Horst DJ (1999) Apolipophorin II/I, apolipoprotein B, vitellogenin, and microsomal triglyceride transfer protein genes are derived from a common ancestor. J Mol Evol 49:150–160. https://doi.org/10.1007/PL00006528
Babin PJ, Carnevali O, Lubzens E, Schneider WJ (2007) Molecular aspects of oocyte vitellogenesis in fish. In: Babin PJ, Cerdà J, Lubzens E (eds) The fish oocyte. Springer, Dordrecht, pp. 39–76. https://doi.org/10.1007/978-1-4020-6235-3_2
Babio L, Lokman PM, Damsteegt EL, Dutoit L (2022) Are cell junctions implicated in the regulation of vitellogenin uptake? insights from an RNAseq-based study in eel. Anguilla Australis Cells 11:550. https://doi.org/10.3390/cells11030550
Bujo H, Hermann M, Kaderli MO, Jacobsen L, Sugawara S, Nimpf J, Yamamoto T, Schneider WJ (1994) Chicken oocyte growth is mediated by an eight ligand binding repeat member of the LDL receptor family. EMBO J 13:5165–5175. https://doi.org/10.1002/j.1460-2075.1994.tb06847.x
Bujo H, Lindstedt KA, Hermann M, Dalmau LM, Nimpf J, Schneider WJ (1995) Chicken oocytes and somatic cells express different splice variants of a multifunctional receptor. J Biol Chem 270:23546–23551. https://doi.org/10.1074/jbc.270.40.23546
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421. https://doi.org/10.1186/1471-2105-10-421
Chen JN, Samadi S, Chen WJ (2015) Elopomorpha (Teleostei) as a new model fish group for evolutionary biology and comparative genomics. In: P Pontarotti (ed) Evolutionary biology: biodiversification from genotype to phenotype. Springer, Cham, pp. 329–344. https://doi.org/10.1007/978-3-319-19932-0_17
Cuchillo-Ibañez I, Lennol MP, Escamilla S, Mata-Balaguer T, Valverde-Vozmediano L, Lopez-Font I, Ferrero I, Sáez-Valero J (2021) The apolipoprotein receptor LRP3 compromises APP levels. Alzheimer’s Res Ther 13:1–17. https://doi.org/10.1186/s13195-021-00921-5
Damsteegt EL, Mizuta H, Ozaki Y, Hiramatsu N, Todo T, Hara A, Ijiri S, Adachi S, Lokman PM (2014) Development and partial characterisation of an antiserum against apolipoprotein B of the short-finned eel, Anguilla australis. J Com Physiol B 184:589–599. https://doi.org/10.1007/s00360-014-0821-4
Damsteegt EL, Falahatimarvast A, McCormick SPA, Lokman PM (2015a) Triacylglyceride physiology in the short-finned eel, Anguilla australis – changes throughout early oogenesis. Am J Physiol Regul Integr Comp Physiol 308:R935–R944. https://doi.org/10.1152/ajpregu.00436.2014
Damsteegt EL, Mizuta H, Hiramatsu N, Lokman PM (2015b) How do eggs get fat? Insights into ovarian fatty acid accumulation in the shortfinned eel, Anguilla australis. Gen Comp Endocrinol 221:94–100. https://doi.org/10.1016/j.ygcen.2014.12.019
Damsteegt EL, Thomson-Laing G, Wylie MJ, Lokman PM (2020) Effects of estradiol and 11-ketotestosterone pre-treatment on artificial induction of maturation in silver female shortfinned eels (Anguilla australis). Plos One 15:e0229391. https://doi.org/10.1371/journal.pone.0229391
Davail B, Pakdel F, Bujo H, Perazzolo LM, Waclawek M, Schneider WJ, Le Menn F (1998) Evolution of oogenesis: the receptor for vitellogenin from the rainbow trout. J Lipid Res 39:1929–1937. https://doi.org/10.1016/S0022-2275(20)32491-3
Dieckmann M, Dietrich MF, Herz J (2010) Lipoprotein receptors - an evolutionarily ancient multifunctional receptor family. Biol Chem 391:1341–1366. https://doi.org/10.1515/bc.2010.129
Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW InfoStat version 2018. InfoStat Group, Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, Argentina. http://www.infostat.com.ar
Dominguez GA, Quattro JM, Denslow ND, Kroll KJ, Prucha MS, Porak WF, Grier HJ, Sabo-Attwood TL (2012) Identification and transcriptional modulation of the largemouth bass, Micropterus salmoides, vitellogenin receptor during oocyte development by insulin and sex steroids. Biol Reprod 87:67–71. https://doi.org/10.1095/biolreprod.112.099812
Endo T, Todo T, Lokman PM, Kudo H, Ijiri S, Adachi S, Yamauchi K (2011) Androgens and very low density lipoprotein are essential for the growth of previtellogenic oocytes from Japanese eel, Anguilla japonica, in vitro. Biol Reprod 84:816–825. https://doi.org/10.1095/biolreprod.110.087163
Esser V, Limbird LE, Brown MS, Goldstein JL, Russell DW (1988) Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J Biol Chem 263:13282–13290. https://doi.org/10.1016/S0021-9258(18)37702-0
Falahati A, Ozaki Y, Damsteegt EL, Zadmajid V, Freeman KJ, Lokman PM (2021) Spatiotemporal expression of activin receptor-like kinase-5 and bone morphogenetic protein receptor type II in the ovary of shortfinned eel, Anguilla australis. Comp Biochem Physiol B Biochem Mol Biol 251:110509. https://doi.org/10.1016/j.cbpb.2020.110509
Gan S, Ye J, Li J, Hu C, Wang J, Xu D, Pan X, Chu C, Chu J, Zhang J, Zheng J, Zhang X, Xu J, Zhang H, Qu F, Cui X (2020) LRP11 activates β-catenin to induce PD-L1 expression in prostate cancer. J Drug Target 28:508–515. https://doi.org/10.1080/1061186X.2019.1687710
Gao XM, Zhang DD, Hou CC, Du C, Luo SY, Zhu JQ (2020) Developmental and mRNA transcript relative abundance pattern of vitellogenin receptors, LR8-/Lrp13, during ovarian development in the large yellow croaker (Larimichthys crocea). Anim Reprod Sci 213:106271. https://doi.org/10.1016/j.anireprosci.2019.106271
Gasteiger, E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook., Springer Protocols Handbook, Humana Press, Totowa, pp 571–607. https://doi.org/10.1385/1-59259-890-0:571
Go GW, Mani A (2012) Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J Biol Med 85:19
Gui Y, Duan Z, Qiu X, Tang W, Gober HJ, Li D, Wang L (2016) Multifarious effects of 17-β-estradiol on apolipoprotein E receptors gene expression during osteoblast differentiation in vitro. BioSci Trends 10:54–66. https://doi.org/10.5582/bst.2016.01006
Harwood BN, Cross SK, Radford EE, Haac BE, De Vries WN (2008) Members of the WNT signaling pathways are widely expressed in mouse ovaries, oocytes, and cleavage stage embryos. Dev Dyn 237:1099–1111. https://doi.org/10.1002/dvdy.21491
Hiramatsu N, Chapman RW, Lindzey JK, Haynes MR, Sullivan CV (2004) Molecular characterization and expression of vitellogenin receptor from white perch (Morone americana). Biol Reprod 70:1720–1730. https://doi.org/10.1095/biolreprod.103.023655
Hiramatsu N, Luo W, Reading BJ, Sullivan CV, Mizuta H, Ryu Y-W, Nishimiya O, Todo T (2013) Hara, A. Multiple ovarian lipoprotein receptors in teleosts. Fish Physiol Biochem 39:29–32. https://doi.org/10.1007/s10695-012-9612-6
Hiramatsu N, Todo T, Sullivan CV, Schilling J, Reading BJ, Matsubara T, Ryu Y-W, Mizuta H, Luo W, Nishimiya O, Wu M, Mushirobira Y, Yilmaz O, Hara A (2015) Ovarian yolk formation in fishes: molecular mechanisms underlying formation of lipid droplets and vitellogenin-derived yolk proteins. Gen Comp Endocrinol 221:9–15. https://doi.org/10.1016/j.ygcen.2015.01.025
Iijima H, Miyazawa M, Sakai J, Magoori K, Ito MR, Suzuki H, Nose M, Kawarabayasi Y, Yamamoto TT (1998) Expression and characterization of a very low density lipoprotein receptor variant lacking the O-linked sugar region generated by alternative splicing. J Biochem 124:747–755. https://doi.org/10.1093/oxfordjournals.jbchem.a022175
Jéhannet P, Kruijt L, Damsteegt EL, Swinkels W, Heinsbroek LT, Lokman PM, Palstra AP (2019) A mechanistic model for studying the initiation of anguillid vitellogenesis by comparing the European eel (Anguilla anguilla) and the shortfinned eel (A. australis). Gen Comp Endocrinol 279:129–138. https://doi.org/10.1016/j.ygcen.2019.02.018
Jonas A, Phillips MC (2008) Lipoprotein structure. In: Vance DE, Vance JE (eds) Biochemistry of lipids, lipoproteins and membranes, 5th edn. Elsevier, pp. 485–506. https://doi.org/10.1016/B978-044453219-0.50019-2
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. https://doi.org/10.1093/bioinformatics/btm091
Kozarsky K, Kingsley D, Krieger M (1988) Use of a mutant cell line to study the kinetics and function of O-linked glycosylation of low density lipoprotein receptors. Proc Natl Acad Sci 85:4335–4339. https://doi.org/10.1073/pnas.85.12.4335
Kroupova H, Trubiroha A, Wuertz S, Kloas W (2011) Stage-dependent differences in RNA composition and content affect the outcome of expression profiling in roach (Rutilus rutilus) ovary. Comp Biochem Physiol Part A Mol Integr Physiol 159:141–149. https://doi.org/10.1016/j.cbpa.2011.02.007
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:458–460. https://doi.org/10.1093/nar/gkaa937
Levene H (1960) Robust tests for equality of variances. In: Olkins I (ed) Contributions to probability and statistics. Stanford University Press, California, pp 278–292
Li Y, Cam J, Bu G (2001) Low-density lipoprotein receptor family. Mol Neurobiol 23:53–67. https://doi.org/10.1385/MN:23:1:53
Li A, Sadasivam M, Ding JL (2003) Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and apolipoprotein B/E: the first three ligand-binding repeats of VtgR interact with the amino-terminal region of Vtg. J Biol Chem 278:2799–2806. https://doi.org/10.1074/jbc.M205067200
Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK (2008) LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev 88:887–918. https://doi.org/10.1152/physrev.00033.2007
Lokman PM, Kazeto Y, Ijiri S, Young G, Miura T, Adachi S, Yamauchi K (2003) Ovarian mitochondrial cytochrome b mRNA levels increase with sexual maturity in freshwater eels (Anguilla spp.). J Comp Physiol B 173:11–19. https://doi.org/10.1007/s00360-002-0304-x
Lokman PM, George KAN, Divers SL, Algie M, Young G (2007) 11-Ketotestosterone and IGF-I increase the size of previtellogenic oocytes from shortfinned eel, Anguilla australis, in vitro. Reproduction 133:955–967. https://doi.org/10.1530/REP-06-0229
Lokman PM, Wylie MJ, Downes M, Di Biase A, Damsteegt EL (2015) Artificial induction of maturation in female silver eels, Anguilla australis: The benefits of androgen pre-treatment. Aquaculture 437:111–119. https://doi.org/10.1016/j.aquaculture.2014.11.026
Lokman PM, Damsteegt EL, Wallace J, Downes M, Goodwin SL, Facoory LJ, Wylie MJ (2016) Dose-responses of male silver eels, Anguilla australis, to human chorionic gonadotropin and 11-ketotestosterone in vivo. Aquaculture 463:97–105. https://doi.org/10.1016/j.aquaculture.2016.05.009
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song SJ, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng D, Lanczycki CJ, Marchler-Bauer A (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. https://doi.org/10.1093/nar/gkz991
Lubzens E, Bobe J, Young G, Sullivan CV (2017) Maternal investment in fish oocytes and eggs: the molecular cargo and its contributions to fertility and early development. Aquaculture 472:107–143. https://doi.org/10.1016/j.aquaculture.2016.10.029
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey A, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. https://doi.org/10.1093/nar/gkz268
Maglott D, Ostell J, Pruitt KD, Tatusova T (2011) Entrez gene: gene-centered information at NCBI. Nucleic Acids Res 39:52–57. https://doi.org/10.1093/nar/gkq1237
Magrané J, Casaroli-Marano RP, Reina M, Gåfvels M, Vilaró S (1999) The role of O-linked sugars in determining the very low density lipoprotein receptor stability or release from the cell. FEBS Lett 451:56–62. https://doi.org/10.1016/S0014-5793(99)00494-9
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:W327–W331. https://doi.org/10.1093/nar/gkh454
Matz MV, Wright RM, Scott JG (2013) No control genes required: Bayesian analysis of qRT-PCR data. PLoS One 8:e71448. https://doi.org/10.1371/journal.pone.0071448
May P, Bock HH, Nimpf J, Herz J (2003) Differential glycosylation regulates processing of lipoprotein receptors by γ-secretase. J Biol Chem 278:37386–37392. https://doi.org/10.1074/jbc.M305858200
May P, Woldt E, Matz RL, Boucher P (2007) The LDL receptor-related protein (LRP) family: An old family of proteins with new physiological functions. Ann Med 39:219–228. https://doi.org/10.1080/07853890701214881
Mizuta H, Luo W, Ito Y, Mushirobira Y, Todo T, Hara A, Reading BJ, Sullivan CV, Hiramatsu N (2013) Ovarian expression and localization of a vitellogenin receptor with eight ligand binding repeats in the cutthroat trout (Oncorhynchus clarki). Comp Biochem Physiol B Biochem Mol Biol 166:81–90. https://doi.org/10.1016/j.cbpb.2013.07.005
Morini M, Lafont AG, Maugars G, Baloche S, Dufour S, Asturiano JF, Pérez L (2020) Identification and stable expression of vitellogenin receptor through vitellogenesis in the European eel. Anim 14:1213–1222. https://doi.org/10.1017/S1751731119003355
Mushirobira Y, Mizuta H, Luo W, Todo T, Hara A, Reading BJ, Sullivan CV, Hiramatsu N (2015) Molecular cloning and partial characterization of a low-density lipoprotein receptor-related protein 13 (Lrp13) involved in vitellogenin uptake in the cutthroat trout (Oncorhynchus clarki). Mol Reprod Dev 82:986–1000. https://doi.org/10.1002/mrd.22579
NCBI Resource Coordinators (2018) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13. https://doi.org/10.1093/nar/gkx1095
Nguyen AT, Chia JH, Kazeto Y, Wylie MJ, Lokman PM (2020) Induction of oocyte development in previtellogenic eel. Anguilla australis. Gen Comp Endocrinol 291:113404. https://doi.org/10.1016/j.ygcen.2020.113404
Nguyen NTT, Vincens P, Dufayard J-F, Crollius HR, Louis A (2022) Genomicus in 2022: comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res 50:D1025–D1031. Available online: https://www.genomicus.bio.ens.psl.eu/genomicus-104.02/cgi-bin/search.pl (Accessed on 1 October 2021).
Nicol B, Guiguen Y (2011) Expression profiling of Wnt signaling genes during gonadal differentiation and gametogenesis in rainbow trout. Sex Dev 5:318–329. https://doi.org/10.1159/000334515
Okabayashi K, Shoji H, Nakamura T, Hashimoto O, Asashima M, Sugino H (1996) cDNA cloning and expression of the Xenopus laevis vitellogenin receptor. Biochem Biophys Res Commun 224:406–413. https://doi.org/10.1006/bbrc.1996.1040
Pohlkamp T, Wasser CR, Herz J (2017) Functional roles of the interaction of APP and lipoprotein receptors. Front Mol Neurosci 10:54. https://doi.org/10.3389/fnmol.2017.00054
Pousis C, Rodríguez C, De Ruvo P, De Virgilio C, Pérez JA, Mylonas CC, Zupa R, Passantino L, Santamaria N, Valentini L, Corriero A (2019) Vitellogenin receptor and fatty acid profiles of individual lipid classes of oocytes from wild and captive-reared greater amberjack (Seriola dumerili) during the reproductive cycle. Theriogenology 140:73–83. https://doi.org/10.1016/j.theriogenology.2019.08.014
Prat F, Coward K, Sumpter JP, Tyler CR (1998) Molecular characterization and expression of two ovarian lipoprotein receptors in the rainbow trout, Oncorhynchus mykiss. Biol Reprod 58:1146–1153. https://doi.org/10.1095/biolreprod58.5.1146
Príncipe C, Dionísio de Sousa IJ, Prazeres H, Soares P, Lima RT (2021) LRP1B: a giant lost in cancer translation. Pharmaceuticals 14:836. https://doi.org/10.3390/ph14090836
Rahman MM, Govindarajulu Z (1997) A modification of the test of Shapiro and Wilk for normality. J Appl Stat 24:219–236. https://doi.org/10.1080/02664769723828
Reading BJ, Hiramatsu N, Sullivan CV (2011) Disparate binding of three types of vitellogenin to multiple forms of vitellogenin receptor in white perch. Biol Reprod 84:392–399. https://doi.org/10.1095/biolreprod.110.087981
Reading BJ, Hiramatsu N, Schilling J, Molloy KT, Glassbrook N, Mizuta H, Luo W, Baltzegar DA, Williams VN, Todo T, Hara A (2014) Lrp13 is a novel vertebrate lipoprotein receptor that binds vitellogenins in teleost fishes [S]. J Lipid Res 55:2287–2295. https://doi.org/10.1194/jlr.M050286
Ren Q, Chen J, Liu Y (2021) LRP5 and LRP6 in Wnt signaling: similarity and divergence. Front. Cell Dev Biol 9:670960. https://doi.org/10.3389/fcell.2021.670960
Scheffé H (1953) A method for judging all contrasts in the analysis of variance. Biometrika 40:87–110. https://doi.org/10.1093/biomet/56.1.229
Schneider WJ (2008) Low-density lipoprotein receptor (LDLR) family: genetics and evolution. eLS. https://doi.org/10.1002/9780470015902.a0006138
Schneider WJ, Nimpf J (2003) LDL receptor relatives at the crossroad of endocytosis and signaling. Cell Mol Life Sci 60:892–903. https://doi.org/10.1007/s00018-003-2183-Z
Schneider WJ, Nimpf J, Brandes C, Drexler M (1999) The low-density lipoprotein receptor family: genetics, function, and evolution. Curr Atheroscler Rep 1:115–122. https://doi.org/10.1007/s11883-999-0007-9
Setiawan AN, Lokman PM (2010) The use of reference gene selection programs to study the silvering transformation in a freshwater eel Anguilla australis: a cautionary tale. BMC Mol Biol 11:1471–2199. https://doi.org/10.1186/1471-2199-11-75
Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Steyrer E, Barber DL, Schneider WJ (1990) Evolution of lipoprotein receptors. The chicken oocyte receptor for very low density lipoprotein and vitellogenin binds the mammalian ligand apolipoprotein E. J Biol Chem 265:19575–19581. https://doi.org/10.1016/S0021-9258(17)45410-X
Stifani S, Barber DL, Nimpf J, Schneider WJ (1990) A single chicken oocyte plasma membrane protein mediates uptake of very low density lipoprotein and vitellogenin. Proc Natl Acad Sci USA 87:1955–1959. https://doi.org/10.1073/pnas.87.5.1955
Taira K, Bujo H, Hirayama S, Yamazaki H, Kanaki T, Takahashi K, Ishii I, Miida T, Schneider WJ, Saito Y (2001) LR11, a mosaic LDL receptor family member, mediates the uptake of ApoE-rich lipoproteins in vitro. Arterioscler Thromb Vas Biol 21:1501–1506. https://doi.org/10.1161/hq0901.094500
Takezaki N (2021) Resolving the early divergence pattern of teleost fish using genome-scale data. Genome Biol Evol 13:evab052. https://doi.org/10.1093/gbe/evab052
Tevosian SG, Manuylov NL (2008) To β or not to β: Canonical β-catenin signaling pathway and ovarian development. Dev Dyn 237:3672–3680. https://doi.org/10.1002/dvdy.21784
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3 - new capabilities and interfaces. Nucleic Acids Res 40:e115. https://doi.org/10.1093/nar/gks596
Wang N, Wang R, Hu Q, Xu W, Zhu Y, Yan F, Chen S (2017) Characterization of a low-density lipoprotein receptor, Lrp13, in Chinese tongue sole (Cynoglossus semilaevis) and medaka (Oryzias latipes). Fish Physiol Biochem 43:1289–1298. https://doi.org/10.1007/s10695-017-0372-1
Wang Y, Han S, You X, Shi X, Liu L, Sun Y, Ma Y, Qian Q, Liu H, Cui B, Zhang Y (2019) The role of low density lipoprotein receptor-related protein 11 as a tumor promoter in cervical cancer. Cancer Manag Res 11:8081. https://doi.org/10.2147/CMAR.S211912
Wasser CR, Masiulis I, Durakoglugil MS, Lane-Donovan C, Xian X, Beffert U, Agarwala A, Hammer RE, Herz J (2014) Differential splicing and glycosylation of Apoer2 alters synaptic plasticity and fear learning. Sci Signal 7:ra113–ra113. https://doi.org/10.1126/scisignal.2005438
Webb JC, Patel DD, Knight JMD, BL. Soutar AK, (1994) Characterization and tissue-specific expression of the human ‘very low density lipoprotein (VLDL) receptor’ mRNA. Hum Mol Genet 3:531–537. https://doi.org/10.1093/hmg/3.4.531
Xu J, Cai R, Lu L, Duan C, Tao X, Chen D, Liu Y, Wang X, Cao M, Chen Y (2014) Genetic regulatory network analysis reveals that low density lipoprotein receptor-related protein 11 is involved in stress responses in mice. Psychiatry Res 220:1131–1137. https://doi.org/10.1016/j.psychres.2014.09.002
Ye ZJ, Go GW, Singh R, Liu W, Keramati AR, Mani A (2012) LRP6 protein regulates low density lipoprotein (LDL) receptor-mediated LDL uptake. J Biol Chem 287:1335–1344. https://doi.org/10.1074/jbc.M111.295287
Zhao N, Liu CC, Qiao W, Bu G (2018) Apolipoprotein E, receptors, and modulation of Alzheimer’s disease. Biol Psychiatry 83:347–357. https://doi.org/10.1016/j.biopsych.2017.03.003
Zheng P, Vassena R, Latham K (2006) Expression and downregulation of WNT signaling pathway genes in rhesus monkey oocytes and embryos. Mol Reprod Dev 73:667–677. https://doi.org/10.1002/mrd.20428
Acknowledgements
The authors would like to thank Dr Martyn Kennedy (Department of Zoology, University of Otago) for his constructive comments around the phylogenetic analysis.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by funding from a University of Otago Doctoral Scholarship (to LB) and from a Research Enhancement Grant (to PML).
Author information
Authors and Affiliations
Contributions
Conceptualisation, L.B.; Methodology, L.B.; Formal analysis, L.B.; Investigation*, L.B.; Project administration, L.B.; Resources, L.B, P.M.L; Supervision, P.M.L., E.L.D.; Validation, L.B., P.M.L., E.L.D.; Visualisation, L.B.; Writing – original draft –, L.B.; Writing – review and editing –, L.B., P.M.L., E.L.D. All authors read and approved the final manuscript. *The ovarian artificial maturation experiment was conducted in 2017 by Damsteegt et al. (unpublished data) and Mercuriali et al. (unpublished data).
Corresponding author
Ethics declarations
Ethical Approval
Fish handling during the ovarian artificial maturation experiment (Damsteegt et al. unpublished data; Mercuriali et al. unpublished data) was approved by the University of Otago Animal Ethics Committee and conducted in accordance with the guidelines of the Australian and New Zealand Council for the Care of Animals in Research and Teaching.
Consent for Publication
Not applicable.
Consent to Participate
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Babio, L., Damsteegt, E.L. & Lokman, P.M. Lipoprotein receptors in ovary of eel, Anguilla australis: molecular characterisation of putative vitellogenin receptors. Fish Physiol Biochem 49, 117–137 (2023). https://doi.org/10.1007/s10695-023-01169-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-023-01169-6