Abstract
Uveal melanoma (UM) is a rare tumor originating from melanocytic cells in the eye. Familial aggregation of UM is rare and can occur as part of the tumor predisposition syndrome BAP1-TPDS. However, family history alone will only identify a subset of patients with BAP1-TPDS. In the present study, we used sequential testing of tumor and blood DNA from UM patients for differential diagnosis of BAP1-TPDS. The study group was an unselected prospective cohort of patients from whom UM tissue was available. First, chromosome 3 status in tumor DNA was determined in all 140 patients who consented to participate. As tumors with disomy 3 rarely show BAP1 alterations, sequence analysis of this gene was performed in the 72 tumors with monosomy 3 (M3) or partial M3 only. We identified oncogenic BAP1 alterations in 52 of these tumors (72%). Targeted sequencing of DNA from matched peripheral blood showed pathogenic variants in two patients (3.8%) thus proving BAP1-TPDS. Only one of these two patients also had a medical history suggestive of this syndrome. Conversely, in three patients known to have had additional tumors before diagnosis of UM, constitutional heterozygosity for a BAP1 mutation was excluded. Altogether, in 50 patients we could exclude BAP1-TPDS with high diagnostic certainty. The results of our study support that genetic testing for BAP1-TPDS should be offered to all patients with UM. Moreover, as genetic information from the tumor can help exclude heritable risk, the strategy for analysis should include efforts to obtain tumor samples for testing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Uveal melanoma (UM) is a malignant tumor that originates from melanocytic cells in the iris, ciliary body, and choroid of the eye [1]. UM is a rare tumor [1]—and, consequently, average individual risk is low. However, the incidence of UM shows marked differences worldwide with 1.3–8.6 cases per million per year in Europe and 0.2–0.3 cases per million in Africa and Asia [2]. This variation appears to be associated with differences in phenotypic features related to melanocyte functioning such as iris color, skin color, and ability to tan [3]. These phenotypic features are heritable traits, and in fact, some polymorphic genetic variants linked to pigmentation are associated with a risk to UM [4]. However, the strength of these associations is low, with odds ratios typically less than 2 [4]. Consequently, at the level of the individual, the effect of these polymorphic genetic variants on risk to UM has no clinical consequences.
The rare observations of families showing aggregation of patients with UM have suggested that heritable predisposition to UM can be transmitted as an autosomal dominant trait albeit with incomplete penetrance, i.e., absence of phenotype in some carriers [5]. The prototypical example of autosomal dominant tumor predisposition is heritable retinoblastoma [6]. The hypothesis that the development of this tumor depends on a two-step mutation process [7] has guided strategies that led to the identification of the RB1 gene that is responsible for a heritable predisposition to retinoblastoma. Using a similar approach, the BAP1 was identified as a likely candidate gene for a heritable predisposition to UM [8, 9]. The deubiquitinase BAP1 is multifunctional protein with tumor suppressor activity involved in chromatin remodeling, DNA damage response, cell cycle regulation, cell death, and differentiation [10, 11]. It turned out that genetic variants of BAP1 can cause high risk not only to UM but also to a spectrum of other neoplasia [8, 9, 12, 13]. It is well established that heritable risk to UM can be part of a tumor predisposition syndrome with variable manifestation and incomplete penetrance (BAP1-tumor predisposition syndrome, BAP1-TPDS), and the number of families with known oncogenic variants of the BAP1 gene has risen fast [14].
Families suspected to have BAP1-TPDS can benefit from genetic testing. Specifically, if diagnostic testing shows that a patient with UM has a BAP1-TPDS, this justifies repeated surveillance examinations for early detection of metachronous tumors, although details of the monitoring strategy need to be developed [15]. Identification of the family-specific pathogenic BAP1 gene variant facilitates predictive testing in relatives. In cancer predisposition syndromes with incomplete penetrance, relatives heterozygous for the mutation have an increased risk [16]. With predictive genetic testing, it is possible to exclude an increased risk in relatives. In addition to the benefit of psychological relief, overall health care cost is likely to be lower than without genetic testing [16].
Only a small proportion of patients with UM have a family history suggestive of BAP1-TPDS, i.e., most patients have isolated UM. Genetic testing has shown that some of these patients are heterozygous for BAP1 variants that cause BAP1-TPDS [8, 9] and this finding establishes the diagnosis of BAP1-TPDS. However, if the genetic analysis is performed on DNA from blood only and fails to identify a pathogenic BAP1 variant, then a BAP1-TPDS is less likely but not excluded. Because the spectrum of BAP1 variants associated with BAP1-TPDS is very heterogeneous, it has to be expected that the false-negative rate of mutation screening tests is not negligible. In patients with isolated retinoblastoma, this problem is solved by mutation identification on DNA from the tumor first, followed by targeted testing in DNA from blood [17]. With this diagnostic strategy, it is possible to exclude an inherited predisposition in most patients with isolated unilateral retinoblastoma. The goal of the study presented here was to apply this strategy for differential diagnosis of BAP1-TPDS in patients with uveal melanoma.
Material and methods
Patients
We invited all UM patients treated in our clinic between October 2014 and October 2016 and from whom tumor material was available to participate in the study. From these, 140 patients agreed. Tumor samples were obtained either by biopsy sampling, by enucleation or endoresection, depending on treatment of the primary tumor. Chromosome 3 status (disomy 3, or monosomy 3) of all tumor samples was analyzed by microsatellite analysis (MSA). BAP1 sequencing was performed on DNA from all samples with monosomy 3 (M3) or partial losses of chromosome 3 (partM3) (n = 72).
Samples and genetic analysis
Genomic DNA was isolated from enucleated tumors by the phenol–chloroform method as described previously [18]. DNA from biopsy samples was isolated using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). DNA from blood was isolated using the FlexiGene kit (Qiagen). The chromosome 3 status of all tumor samples was determined by STR-genotyping (microsatellite analysis, MSA) using eight chromosome 3 markers as described elsewhere [19]. To detect tumors with isodisomy 3 all tumors showing loss of heterozygosity (LOH) of chromosome 3 markers by MSA were subjected to chromosome 3 MLPA (MRC Holland, probemix P027-C2). BAP1 mutation screening was performed by Sanger sequencing in DNA from 72 UM with M3 or partM3 as described in Martin et al., [20]. In samples without clear results by Sanger sequencing, NGS panel sequencing was performed. Copy number analysis of all BAP1 exons was determined by Multiplex ligation-dependent probe amplification (MLPA) using SALSA MLPA probemix P417-B2 BAP1 (MRC Holland, Amsterdam, The Netherlands) following the manufacturer's instructions.
Panel sequencing
In this study, Agilent SureSelectXT HS target enrichment system was used to enrich the genomic regions of interest. The customized panel was designed to capture all coding regions of BAP1, SF3B1, EIF1AX, CYSLTR2, PCLB4, RB1, and exons 4 and 5 of the GNAQ and GNA11 genes. About 100 ng genomic tumor DNA was subjected to library preparation according to the manufacturer's instructions (protocol version A1, July 2017). Briefly, genomic DNA was fragmented using a Covaris S220 to an average size of 150 to 200 bp, and the fragments were ligated to molecular barcodes. The captured fragments enriched for the target sequences were sequenced on Illumina MiSeq by paired-end sequencing with 2 × 150 bp. We obtained a coverage of > 400 × for all coding regions in each sample. For mapping of the reads and mutation calling, the fastq files were analyzed using SureCall (version 4.1.1.5, Agilent). Sanger sequencing was used to re-sequence regions showing mutations by Panel Sequencing in tumor DNA and to sequence the respective blood DNA of the patient. The pathogenicity of the germline variants was determined using InterVar (https://wintervar.wglab.org/) which is based on the ACMG/AMP 2015 guidelines [21]. For evaluating the oncogenicity of somatic variants we have adhered to the applicable criteria of the ACMG guidelines: Nonsense, frameshift, exon skipping and deletion variants in conjunction with LOH of BAP1 are classified as oncogenic mutations. Missense variations in BAP1 with a ClinVar or COSMIC entry are classified as oncogenic if in addition computational data indicate impaired protein function.
Results
Participants
All patients (n = 265) who requested prognostic biomarker testing (chromosome 3 status in tumor DNA) were eligible (148 males (55.8%), median age = 61.3 years, interquartile range (IQR) = 15.8 years and 117 female, median age = 61.7 years, IQR = 21.2 years). Of these, 140 (52.8%) consented to participate in this study (83 males (59.3%) with median age at diagnosis = 60.8 years and IQR = 14 years; 57 females with median age at diagnosis = 57.9 years and IQR = 20.1 years).
Chromosome 3 status
Mutational inactivation of the BAP1 gene is rarely seen in the class of UMs with disomy 3 (D3) if isodisomy 3 is excluded [22, 23]. To focus the analytical efforts on samples likely to have BAP1 gene alterations we determined the chromosome 3 status in tumors before further mutation analysis. Genotype data of polymorphic STR-loci from 134 patients met our technical quality criteria for determining the chromosome 3 status. These criteria require that a minimum number of loci are heterozygous in DNA from blood. Uveal melanomas from 62 patients (46%) showed no loss of heterozygosity at any informative locus and, therefore, were classified as disomy 3 tumors (Fig. 1). In tumors from 61 patients (46%), monosomy 3 was diagnosed because of loss of heterozygosity at all informative loci (Fig. 1). In UMs from the remaining 11 patients (6%), only some of the informative loci on chromosome 3 showed allele loss (Fig. 1). This pattern of allele loss is typical for uveal melanomas with deletions only of parts of chromosome 3 (partM3).
Mutation analysis of the BAP1 gene in uveal melanoma
BAP1 mutation analysis was performed in UM with monosomy 3 or partial deletions of chromosome 3 (72 tumors). Sanger sequencing was chosen as the first-line method of sequencing analysis. Those samples that did not meet the quality criteria for mutation analysis, which require, among other parameters, that sequence data has been obtained for all coding and invariant splice site regions of the BAP1 gene, were subjected to panel sequencing which included the BAP1 gene. Copy number variant detection by MLPA was performed on samples that did not show a oncogenic small variant. Alterations of the BAP1 gene were identified in 52 of the 72 tumors with M3 or partM3 (72%). In the tumors with partM3 the percentage of samples with BAP1 mutation was lower (27%) than in the M3 group.
In 50 of 52 tumors we detected oncogenic BAP1 alterations (Table 1). Alterations identified in these 50 tumors included two deletions that spanned one or more exons. The remaining 48 alterations were small variants. In these tumors the signal of the normal allele was low compared to that of the variant allele. This result is in line with hemizygosity at the BAP1 locus, which is expected in tumors with monosomy 3. In tumors with alterations only of parts of chromosome 3 this result suggests that the BAP1 locus is within regions of allele loss.
Two of 52 tumors did not show a bona fide pathogenic alteration but an identical single base substitution at the -4 position of the splice donor site of exon 17. This position is not part of the invariant splice donor sequence motive and the base substitution does not create an alternative donor site. In silico analysis of this intronic variant have previously shown that it is not expected to affect splicing [24]. In both patients, this variant allele was also present in the constitutional DNA and, therefore, it is plausible that this alteration is a rare and likely neutral polymorphic variant.
Analysis of DNA from blood for patient-specific variant BAP1 alleles
We performed BAP1 mutation analysis targeting the oncogenic variants detected in the tumor on DNA from matched peripheral blood (50 patients). In 48 patients (96%) results showed that the oncogenic variant identified in the tumor resulted from a somatic mutation in the patient. Two patients (4%) showed heterozygosity for the oncogenic variant identified in the tumor, thus indicating a germ-line origin (TP7, TP24). Both variants were classified as pathogenic by “InterVar” based on ACMG criteria. One of the germline mutations p.(Ser126Glnfs*16) has been identified in a patient with mesothelioma, previously [25]. As we had no access to constitutional DNA from parents or other relatives of these two patients, we could not track the origin of the variants any further.
Functional types and localization of oncogenic alterations
Missense and in-frame length variants (11 and 5, respectively) were identified in samples from 16 patients, and all were of somatic origin (Table 1). Amino acids predicted to be altered by these mutations were all located in the N-terminal UCH domain of the BAP1 protein (Fig. 2a). One missense variant, c.188C > G [p.(Ser63Cys)], was identified in tumors of two patients and is also listed as a recurrent somatic variant (n = 5) in the Catalogue Of Somatic Mutations In Cancer (COSMIC V90_38_MUTANTCENSUS). Of the other 14 variants predicted to alter only single or few amino acids of the BAP1 gene 9 variants (56%) are also listed in COSMIC. The interpretation of one missense mutation p.(Ser98Arg) although located in the UCH domain and present on the background of chromosome 3 loss remains uncertain as computational data do not indicate a severe impairment of protein function.
Map of BAP1 oncogenic sequence variants in 50 uveal melanomas. a Black triangles: location of somatic missense or in-frame variants. b and c Black circles: location of premature stop codons due to nonsense, frameshift or splice variants caused by somatic alterations. Red circles: location of premature stop codons due to nonsense, frameshift or splice variants caused by constitutional alterations. Grey bar: in-frame loss of codons due to splice variant caused by somatic alteration.
Tumors from 32 patients had variants resulting in premature termination codons (3 nonsense, 19 frame-shift, and ten splice site variants; Table 1). The locations of these terminations showed no clustering to particular domains of the BAP1 protein (Fig. 2b). No premature termination was located within the last exon and the 50 bp 3′-parts of the penultimate exon of the BAP1, which are regions expected not to elicit nonsense-mediated decay [26]. One of the variants resulting in premature termination codons, c.588G > A [p.(Trp196Ter)], was identified in tumors of two patients and is also listed as a recurrent somatic variant (n = 4) in COSMIC. However, in all, only 4 of the 28 (14%) premature termination variants identified here are also listed in COSMIC.
One tumor (TP1) showed a deletion of the splice acceptor-site of intron 5 and parts of exon 5 (Table 1). The expected consequence of this variant is a loss of exon 5 from the mRNA and, on translation, an in-frame deletion of Leu86 (CTG) to Glu125(GAG) (Fig. 2c). These amino acids are part of the N-terminal UCH domain of the BAP1 protein.
Relations between constitutional BAP1 genotype and phenotypic features
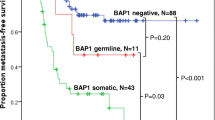
The ages at diagnosis of the two patients with pathogenic BAP1 germ-line variants ranked at positions 4 and 21 within the group of 50 patients with BAP1 variants. Their age rank positions within the whole group of 140 patients were similar (positions 12 and 71 of 140). In the density plots of age at diagnosis, the two patients with germ-line variants also do not appear to be outliers (Fig. 3).
At the time of diagnosis of the primary tumor, data on medical history concerning tumors outside of the eye was available from 100 patients. Two patients had a history of renal cell cancer diagnosed prior to uveal melanoma but only one of these two patients was heterozygous for a pathogenic BAP1 germ-line mutation. This patient also had a history of cutaneous malignant melanoma. In the two other patients with diagnosis of cutaneous malignant melanoma prior to uveal melanoma, a common pathogenic BAP1 gene germline variant as root cause is highly unlikely because the BAP1 gene mutations that we identified in their tumors were not present in DNA from blood and, therefore, of somatic origin. However, as we cannot exclude low-dose mosaicism and thus it is conceivable that tumors can originate from cells of a mutant sector that share a oncogenic BAP1 gene somatic variant. Data on family history was incomplete for most patients. Of note, the father of a BAP1 heterozygous patient in our study had died from pleural mesothelioma, a tumor type that is very rare in the general population but relatively frequent in families with BAP1-TPDS (for review see [14]).
Discussion
In this study we used genetic testing of tumors for molecular differential diagnosis of BAP1-TPDS in patients with UM. We adapted a stepwise testing strategy similar to the one that is used for routine in genetic testing in retinoblastoma to the requirements specific for uveal melanoma. The initial analysis step identifies tumors with complete or partial monosomy 3 and, in the next step, DNA from these tumors is analyzed for BAP1 gene alterations. With decreasing sequencing costs due to NGS, we would suggest sequencing of tumor and blood tissue simultaneously instead of one after another which is more time efficient. A BAP1-TPDS is excluded in patients in whom a somatic origin of these alterations is established by analysis of constitutional DNA. With this three-step strategy, we could safely exclude a BAP1-TPDS in 50 of 72 (70%) patients with a M3 or partM3 UM. Since germ-line variant alleles of the BAP1 gene are rare in UM patients with D3 tumors (odds ratio 0.12, derived from data published by Ewens 2018 [22]), TPDS is also unlikely in the group of 62 patients with D3 tumors. However, it remains to be shown if extending the diagnostic strategy to patients with UM with D3 leads to detection of further BAP1-TPDS with a frequency higher than those in the normal (UM-free) population [22].
Most of the alterations with confirmed somatic origin resulted in premature termination outside of the regions that are expected to evade nonsense mediated mRNA decay [26] and, therefore, are expected to result in loss of function, which is typical for the spectrum of BAP1 alterations in cancer. All amino acids altered by missense substitution or short in-frame losses belonged to the ubiquitin C-terminal hydrolase (UCH) domain of the BAP1 protein. This catalytic domain plays a key role by mediating deubiquitination of chromatin elements such as H2A [27]. Most of the missense substitutions identified here have been reported as somatic mutations in other tumors and this also supports that these alterations can be classified as pathogenic. Recurrence of somatic missense mutations was also found in a recent report by Ewens et al. [22]. Thus, it appears that the set of oncogenic missense mutations of the BAP1 gene is smaller than the set of variants that result in premature termination.
We did not identify oncogenic BAP1 alterations in DNA from 14 to 8 tumors (29%) with M3 or partM3, respectively. In the tumors with partM3 the percentage of samples with BAP1 mutation was much lower (27%) compared to M3 tumors which is in concordance with the observation that most of these tumors show a mutation pattern typical for the UM with D3 [20]. Between reported studies the proportion of tumors with M3 with a oncogenic BAP1 alteration varies and the detection rate in our study is well within this range [23, 28]. It is conceivable that some tumors with M3 may have BAP1 gene alterations that are not within the scope of the detection methods used here. Specifically, we expect an improved rate of detection if sequencing libraries cover the complete BAP1 gene region instead of focusing enrichment of the exons only. The need for extension of the scope of analysis by NGS has already been pointed out by Field et al. 2018 [23, 28]. Another advantage of NGS-based analysis is a lower detection limit compared to Sanger sequencing. This allows the detection of sequence variants in heterogeneous tumor tissue containing high proportion of normal cells [23] thus increasing the proportion of fully informative cases, although it does not lead to the discovery of additional heritable cases.
In 2 patients (4% of 50 patients with BAP1 mutant tumors, 1.4% of all 140 patients) we determined heterozygosity for pathogenic BAP1 gene variants and this established the diagnosis of a BAP1-TPDS. Both variants are expected to result in premature termination which is the predominant type of consequence of pathogenic BAP1 alterations. Premature termination was also the most frequent biological consequence among the BAP1 germ-line variants reported by Ewens et al. [22]. By contrast, studies restricted to mutation analysis on DNA from blood tend to report a higher proportion of missense type variants and these include a high proportion of variants of uncertain pathogenic significance (VUS) [22]. In fact, 608 of 738 (82%) gene variants in the BAP1 coding region listed in ClinVar in the context of heritable tumor susceptibility are VUS (https://www.ncbi.nlm.nih.gov/clinvar, query of 14.7.2021). Using our strategy, which requires initial sequencing of tumor DNA, only half as much VUS are expected due to loss of one BAP1 allele in most tumors. In other words, results of mutation analysis on DNA from blood only are often insufficient to provide the evidence needed to demonstrate or exclude a BAP1-TPDS with high confidence. In vitro assays that assess if some functions of BAP1 protein are altered depending on certain alterations can provide additional data in support of the pathogenicity of exonic BAP1 variants [29, 30]. However, we agree that “further research into BAP1 functions is needed” to arrive at “a consensus on mandatory experimental assays to assess VUS” [30].
Only one of the two patients that we identified to be heterozygous for a pathogenic BAP1 alteration was also positive for features proposed as referral guidelines for genetic diagnostics of a BAP1-TPDS [31]. This patient was diagnosed with CMM and RCC (at age 28 and 39 years, respectively) prior to the diagnosis of UM (at age 59 years). Moreover, one parent of this patient had died of pleural mesothelioma, a rare cancer that is frequent in patients with a BAP1-TPDS. The medical and family history of the other BAP1-TPDS patient identified in our study did not meet the referral criteria proposed by Chau et al. [31]. However, with a relatively young age at diagnosis of UM (40 years) this patient qualifies according to more relaxed guidelines [9]. From recent studies it appears that population-based analyses, which avoid the ascertainment biases introduced by selection of high-risk patients, are required to obtain a better understanding of the BAP1-TPDS [22]. In other cancer predisposition syndromes such as hereditable breast and ovarian cancer (HBOC) it has turned out that initial estimates of penetrance, which were derived from observations of familial aggregation of cancer cases, were inflated. Unbiased data are required to model the cancer risk of individuals who are heterozygous for a pathogenic BAP1 allele. Such a statistical model is essential to derive surveillance plans for early detection. The results of our study support that genetic diagnosis of a BAP1-TPDS should be offered to all patients with UM. Moreover, the testing strategy should include efforts to obtain genetic information from the tumor.
Data availability
Not applicable.
Code availability
Not applicable.
References
Kaliki S, Shields CL (2017) Uveal melanoma: relatively rare but deadly cancer. Eye (Lond) 31(2):241–257. https://doi.org/10.1038/eye.2016.275
Krantz BA, Dave N, Komatsubara KM, Marr BP, Carvajal RD (2017) Uveal melanoma: epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol 11:279–289. https://doi.org/10.2147/OPTH.S89591
Weis E, Shah CP, Lajous M, Shields JA, Shields CL (2006) The association between host susceptibility factors and uveal melanoma: a meta-analysis. Arch Ophthalmol 124(1):54–60. https://doi.org/10.1001/archopht.124.1.54
Ferguson R, Vogelsang M, Ucisik-Akkaya E et al (2016) Genetic markers of pigmentation are novel risk loci for uveal melanoma. Sci Rep 6:31191. https://doi.org/10.1038/srep31191
Lynch HT, Krush AJ (1968) Heredity and malignant melanoma: implications for early cancer detection. Can Med Assoc J 99(1):17–21
Vogel F (1954) Genetics and mutation rate of retinoblastoma (glioma retinae), with general remarks on methods of determining mutation rate in humans. Z Mensch Vererb Konstitutionsl 32(4):308–336
Knudson AG Jr (1971) Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 68(4):820–823. https://doi.org/10.1073/pnas.68.4.820
Wiesner T, Obenauf AC, Murali R et al (2011) Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 43(10):1018–1021. https://doi.org/10.1038/ng.910
Abdel-Rahman MH, Pilarski R, Cebulla CM et al (2011) Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet 48(12):856–859. https://doi.org/10.1136/jmedgenet-2011-100156
Han A, Purwin TJ, Aplin AE (2021) Roles of the BAP1 tumor suppressor in cell metabolism. Cancer Res 81(11):2807–2814. https://doi.org/10.1158/0008-5472.CAN-20-3430
Carbone M, Harbour JW, Brugarolas J et al (2020) Biological mechanisms and clinical significance of BAP1 mutations in human cancer. Cancer Discov 10(8):1103–1120. https://doi.org/10.1158/2159-8290.CD-19-1220
Carbone M, Ferris LK, Baumann F et al (2012) BAP1 cancer syndrome: malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med 10:179. https://doi.org/10.1186/1479-5876-10-179
Testa JR, Cheung M, Pei J et al (2011) Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 43(10):1022–1025. https://doi.org/10.1038/ng.912
Walpole S, Pritchard AL, Cebulla CM et al (2018) Comprehensive study of the clinical phenotype of germline BAP1 variant-carrying families worldwide. J Natl Cancer Inst 110(12):1328–1341. https://doi.org/10.1093/jnci/djy171
Star P, Goodwin A, Kapoor R et al (2018) Germline BAP1-positive patients: the dilemmas of cancer surveillance and a proposed interdisciplinary consensus monitoring strategy. Eur J Cancer 92:48–53. https://doi.org/10.1016/j.ejca.2017.12.022
Noorani HZ, Khan HN, Gallie BL, Detsky AS (1996) Cost comparison of molecular versus conventional screening of relatives at risk for retinoblastoma. Am J Hum Genet 59(2):301–307
Lohmann DR, Gerick M, Brandt B et al (1997) Constitutional RB1-gene mutations in patients with isolated unilateral retinoblastoma. Am J Hum Genet 61(2):282–294. https://doi.org/10.1086/514845
Tschentscher F, Husing J, Holter T et al (2003) Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res 63(10):2578–2584
Tschentscher F, Prescher G, Zeschnigk M, Horsthemke B, Lohmann DR (2000) Identification of chromosomes 3, 6, and 8 aberrations in uveal melanoma by microsatellite analysis in comparison to comparative genomic hybridization. Cancer Genet Cytogenet 122(1):13–17. https://doi.org/10.1016/s0165-4608(00)00266-1
Martin M, Masshofer L, Temming P et al (2013) Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 45(8):933–936. https://doi.org/10.1038/ng.2674
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Ewens KG, Lalonde E, Richards-Yutz J, Shields CL, Ganguly A (2018) Comparison of germline versus somatic BAP1 mutations for risk of metastasis in uveal melanoma. BMC Cancer 18(1):1172. https://doi.org/10.1186/s12885-018-5079-x
Field MG, Durante MA, Anbunathan H et al (2018) Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Commun 9(1):116. https://doi.org/10.1038/s41467-017-02428-w
Betti M, Casalone E, Ferrante D et al (2015) Inference on germline BAP1 mutations and asbestos exposure from the analysis of familial and sporadic mesothelioma in a high-risk area. Genes Chromosomes Cancer 54(1):51–62. https://doi.org/10.1002/gcc.22218
Carbone M, Pass HI, Ak G et al (2022) Medical and surgical care of patients with mesothelioma and their relatives carrying germline BAP1 mutations. J Thorac Oncol 17(7):873–889. https://doi.org/10.1016/j.jtho.2022.03.014
Lindeboom RG, Supek F, Lehner B (2016) The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet 48(10):1112–1118. https://doi.org/10.1038/ng.3664
Okino Y, Machida Y, Frankland-Searby S, Machida YJ (2015) BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J Biol Chem 290(3):1580–1591. https://doi.org/10.1074/jbc.M114.609834
van de Nes JA, Nelles J, Kreis S et al (2016) Comparing the prognostic value of BAP1 mutation pattern, chromosome 3 status, and BAP1 immunohistochemistry in uveal melanoma. Am J Surg Pathol 40(6):796–805. https://doi.org/10.1097/PAS.0000000000000645
Hong JH, Chong ST, Lee PH et al (2020) Functional characterisation guides classification of novel BAP1 germline variants. NPJ Genom Med 5:50. https://doi.org/10.1038/s41525-020-00157-6
Repo P, Jarvinen RS, Jantti JE et al (2019) Population-based analysis of BAP1 germline variations in patients with uveal melanoma. Hum Mol Genet 28(14):2415–2426. https://doi.org/10.1093/hmg/ddz076
Chau C, van Doorn R, van Poppelen NM et al (2019) Families with BAP1-tumor predisposition syndrome in The Netherlands: path to identification and a proposal for genetic screening guidelines. Cancers (Basel). https://doi.org/10.3390/cancers11081114
Acknowledgements
We thank Prof. Dr. Hebebrandt for providing the nucleus for the initiation of the study.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by Deutsche Krebshilfe (Grant No.70110960).
Author information
Authors and Affiliations
Contributions
Material preparation, data collection, and analysis were performed by all authors. The first draft of the manuscript was written by Dietmar Lohmann and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. DL: conceptualization; MZ, YAA: methodology; DL: formal analysis and investigation; DL: writing—original draft preparation; MZ: writing—review and editing; DL: funding acquisition.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the University Hospital Essen (13–5462-B0).
Consent to participate
Written informed consent to participate was given by every patient included in this study.
Consent for publication
All patients included in this study signed informed consent regarding publishing their data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abbassi, Y.A., Le Guin, C., Bornfeld, N. et al. Analysis of uveal melanomas and paired constitutional DNA for exclusion of a BAP1-tumor predisposition syndrome. Familial Cancer 22, 193–202 (2023). https://doi.org/10.1007/s10689-022-00310-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-022-00310-3