Abstract
Peutz–Jeghers syndrome (PJS) is a rare hereditary syndrome characterized by the occurrence of hamartomatous polyps in the gastrointestinal tract, mucocutaneous pigmentation and increased risk of cancer in multiple internal organs. PJS is preconditioned by the manifestation of mutations in the STK11 gene. The majority of detected STK11 changes are small scale mutations, however recent studies showed the significant contribution of medium-sized changes commonly known as copy number variations (CNVs). Here we present a novel 7001 bps deletion of STK11 gene fragment, in which we identified the presence of breakpoints (BPs) within the Alu elements. Comparative meta-analysis with the 80 other CNV cases from 12 publications describing STK11 mutations in patients with PJS revealed the participation of specific Alu elements in all deletions of exons 2–3 so far described. Moreover, we have shown their involvement in the two other CNVs, deletion of exon 2 and deletion of exon 1–3 respectively. Deletion of exons 2–3 of the STK11 gene may prove to be the most recurrent large rearrangement causing PJS. In addition, the sequences present in its BPs may be involved in a formation of a significant percentage of the remaining gene CNVs. This gives a new insight into the conditioning of this rare disease and enables improvements in PJS genetic diagnostics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peutz–Jeghers syndrome (PJS; OMIM 175200) is a predisposition to the occurrence of pigment skin lesions and hamartomatous polyps inherited in an autosomal dominant way. The polyps may cause ileus and bleeding from the lower part of the tract, if underwent autoamputation [1, 2]. Incidence of the syndrome ranges from 1/29,000 to 1/120,000 births [3]. Patients with PJS are at an elevated risk of developing extra intestinal cancers located in the pancreas, the breasts, the ovary and the uterus [4–6].
Peutz–Jeghers syndrome is preconditioned by the manifestation of mutations in the STK11 (OMIM*602216 Serine/Threonine Protein Kinase 11) gene, located in the short arm of chromosome 19, in the 13.3 region. The gene consists of ten exons, nine of which code for a protein. The majority of detected changes are small scale mutations, however recent studies showed the contribution of large rearrangements in a significant percentage of patients, reaching in some groups even 30 % of all the detected mutations [7, 8]. These medium-sized changes concerning exons or larger genomic fragments are commonly known as copy number variations (CNV) [9]. Literature indicates a number of mechanisms involved in the formation of CNV [10]. One of the hypotheses described assumes that a significant proportion of large mutations occurs within the interspersed repeats through non-allelic homologous recombination. Alu elements are a family of short interspersed nuclear elements (SINE) and comprise about 10.5 % of the human genome [11]. There is an overrepresentation of such sequences in the STK11 gene [12]. Alu elements, which represent 19 % of the entire gene sequence (24 kb), are concentrated mainly in the distal part of the gene (especially in intron 1). In the following publication we present the case of a patient with a deletion of STK11 gene fragment, in which we observed the presence of breakpoints (BPs) within the Alu elements. We decided to perform a meta-analysis of similar cases reported in the literature, using the in silico analysis of the DNA sequence of the STK11 gene.
Materials and methods
PJS family description
The proband is a 39 year-old woman (born in 1975), who manifested pigmented lesions of the lips and buccal mucosa during infancy. In preschool and school age pigmentation increased, appearing around the mouth, on the eyelids and around fingers. After puberty these signs gradually subsided. Currently 39, she exhibits small muco-cutaneous melanosis around the lips and eyelids. From the age of 21 the patient has undergone surgery four times to remove polyps from the stomach, small intestine and colon, preceded by signs of mechanical obstruction and gastrointestinal bleeding. All removed polyps were hamartomas with no signs of malignant transformation.
The proband’s mother also had pigmented lesions on the lips and buccal mucosa. She underwent surgery twice (in 1985 and 2000) because of gastrointestinal polyps. All observed polyps were also hamartomas with no signs of malignant transformation. The proband’s grandmother died from skin cancer of right groin, diagnosed as a melanoma while her grandfather died aged 54 due to “twisted bowel” (lack of precise clinical data). PJS in the family has been also diagnosed in the proband’s two children, sister and nephew.
Study design
The material for research was DNA from peripheral blood. The blood was collected in cooperation with the Department of Dermatology of Gdansk Medical University. The studies were approved by the local Ethics Committee of the Poznan University of Medical Science and performed after obtaining written informed consent from the patients. The preliminary diagnostics using high resolution melting (HRM) and single strand conformation polymorphism (SSCP) showed no small mutations [13]. Subsequently, the patient was examined for large rearrangements using multiplex ligation-dependent probe amplification (MLPA) method (SALSA MLPA kit P101-A2 [MRC Holland]), which revealed decreased signals for probes hybridizing to exons 2 and 3.
Location of interspersed repeats within the entire sequence (24 kbps) of the STK11 gene was determined on the basis of bioinformatic analysis with the use of RepeatMasker software, version open-3.3.0 (www.repeatmasker.org).
Primer pair encompassing potentially involved Alu sequences within introns 1 and 3 was designed to identify the exact location of mutation breakpoints. Primer sequences were as follows: forward—5′-TTCTGGTAGGCAGTGGGTTC-3′; reverse—5′-TCACAGAAGTCCAGGCACAC-3′. The 8936 bps long PCR product was obtained through Long-range PCR using Advantage Genomic LA Polymerase [Clontech]. Long-range PCR conditions were as follows: 94 °C for 1 min, and then 30 cycles of 94 °C for 30 s, and 68 °C for 13.5 min, and finally 72 °C for 10 min. The 25-μL reaction volume contained 100 ng of DNA and 5 pmol of each primer. PCR products were separated on 1 % agarose gel [Sigma Aldrich] electrophoresis in 1x TBE buffer. The shorter (approximately 2000 bps) product was directly sequenced by Sanger method using ABI 373 DNA sequencer [Applied Biosystems] according to manufacturer’s instructions.
Interspersed repeats alignment was conducted with the use of basic local alignment search tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) optimized for somewhat similar sequences (blastn). Sequence nomenclature used to describe the breakpoints and interspersed repeats positions was based on the UCSC Genome Browser Feb. 2009 (GRCh37/hg19). The nomenclature of all previously described CNVs was converted for the purpose of publication in accordance with the above-mentioned guidelines (Tables 1, 2).
Meta-analysis
We performed a comparative analysis with the 80 CNV cases of the STK11 gene from 12 publications describing STK11 mutations in patients with PJS [7, 8, 12, 14–22].
Results
Breakpoint identification
Using the MLPA and sequencing we identified a 7001 bps deletion c.280 + 5594_464 + 384del7001 co-segregating with PJS phenotype in this family.
The analysis of the STK11 gene sequence using RepeatMasker software revealed the presence of 20 interspersed repeats, including 18 SINE/Alu and 2 LINE1 elements. Comparing the data from bioinformatic analysis of the mutation sequence and the interspersed repeats within the STK11 gene, we found that both mutation breakpoints are located in Alu elements from AluY subfamily, in regions [1,212,558–1,212,842] and [1,219,530–1,219,843] respectively.
In silico analysis results
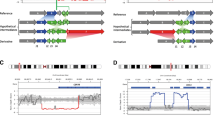
Deletion of exons 2 and 3 of the STK11 gene were observed also in PJS patients from other populations (Table 1). In several studies the exact breakpoint locations of detected CNVs were determined, and the results indicate the involvement of the same regions in the formation of all detected deletions of exons 2 and 3 (Fig. 1).
Approximate location of each STK11 exons 2–3 deletion with identified breakpoint positions. Mutation order corresponds to the description in Table 1
For distal breakpoints (5′) the participating region contains three interspersed repeats next to each other (all from class SINE and Alu family), respectively: AluY comprising sequence [1,212,558–1,212,842] [for the sake of publication it is referred to as AluY(1)], FLAM_C comprising sequence [1,212,850–1,212,978], and second AluY comprising sequence [1,212,891–1,213,195] [referred to as AluY(2)]. All proximal breakpoints (3′) are within the same Alu element from AluY subfamily comprising sequence [1,219,530–1,219,843] [referred to as AluY(3)].
Subsequently, we performed an alignment of AluY(3) sequence with the elements from 5′ BP and all other interspersed repeats within the gene located upstream (5′) from AluY (3). The analysis showed a high complementarity of AluY(3) to AluY(1)—232/295 (79 %), FLAM_C—84/112 (75 %) and AluY(2)—272/305 (89 %) and the position of all the above mentioned elements in the same orientation.
Summary of clinical data
The phenotype of deletion carriers is generally typical for PJS (Table 3). All probands except one (diagnosed at the age of 1) had a characteristic pigmentation and hamartomatous polyps. Polyps in the stomach were observed in 25 % of the cases, and a single proband had nasal polyps. The age of diagnosis is varied. The course of disease in this group is not particularly severe. The available data show that among the probands occurrence of cancer has not been observed. In our proband’s family, melanoma occurred only in her grandmother.
Discussion
Molecular studies of genetic diseases rely on methods able to detect both small sequence variants (point mutations, etc.) and rearrangements of larger genomic fragments. Screening methods, along with direct sequencing, can identify the majority of small mutations. Copy number variations are a greater challenge and the methods used for their detection do not always allow researchers to determine the exact sequence undergoing mutation (e.g. MLPA, Quantitative-PCR, Array-CGH, C-HRM) [23–26]. These approaches not only fail to identify the exact breakpoint but also prevent any further examination of possible mechanisms involved in their formation.
In our study we have detected deletion of exons 2–3 of the STK11 gene in a family with PJS and determined the breakpoints of the mutation. Similar mutations had been described in studies of PJS patients from other populations. Among them 11 were deletions encompassing exons 2 and 3. Exact breakpoints were identified in five cases only, while the remaining ones provided no information on the size and exact location of the change. All six cases described (including our patient) are similar in terms of both BP positions, 3′ BPs in particular, which in all cases are within the AluY(3) element. All distal breakpoints (5′) are also located in the same region, with the difference that in silico analysis revealed the presence of three contiguous interspersed repeats: AluY(1), FLAM_C and AluY(2), respectively. All the above elements belong to the SINE class and Alu family. Although 5′ BPs of changes are not exactly in the same element, it can be assumed that the whole region is involved in the formation of CNV, and the BP position is merely the result of non-specific DNA repair. Involvement of Alu elements in all mutations analyzed may confirm De Rosa’s hypothesis that non-allelic homologous recombination mediated by Alu elements is a factor causing the formation of deletions of exons 2–3 in the STK11 gene [12]. Moreover, this mechanism seems to be recurrent (Fig. 2).
Then we checked the complementarity between AluY(3) and other interspersed repeats present in the gene upstream from AluY(3). Sequence alignment showed high complementarity of AluY(3) to all elements from the distal BPs of analyzed changes. Furthermore, distal as well as proximal BPs are situated in the same orientation, which may be crucial for the mechanism involved in their formation.
Of the 80 CNVs described in publications we analyzed only 20 having determined breakpoints. Excluding five deletions of exons 2–3, BPs in CNVs encompassing this region were determined only for two changes that are: deletion of exon 2 described in Chile and deletion of exons 1–3 in Italy (Table 2). Both these changes share one BP with deletions of exons 2–3. Distal BP of exon 2 deletion is within AluY(2), and proximal BP of exons 1–3 deletion is in AluY(3) (Fig. 3). Of the total 21 CNVs with determined BPs, 8 (over 38 %) exhibit participation of Alu elements from these two regions.
Approximate location of other CNVs with breakpoints present in the Alu elements of interest. Mutation order corresponds to the description in Table 2
Further analysis of the remaining 60 CNVs without specified BPs showed that there are 13 additional changes in which at least one BP is located in intron 1 or 3. It cannot be excluded that in their formation Alu elements of interest are involved. Of the 81 identified CNVs, 8 have confirmed BPs in these regions, while the other 19 are changes with their potential involvement. Hence, there are 27 conceivable changes from 81 CNVs, which represent over 33 % of all detected CNVs of the STK11 gene.
The increased prevalence of CNV in the gene can be associated with the accumulation of interspersed repeats including Alu, especially in the distal region, which has already been observed and thoroughly discussed by Resta et al. [15]. Of the 15 patients with CNV in their group, 7 had deletions caused by NAHR between Alu elements. They defined an 18,019 bp-long region of the STK11 gene as a recombination hotspot. Resta et al. indicated that the repetitive elements have been implicated in chromosomal aberrations because their presence increases instability of the region. Our observations confirm this thesis, furthermore narrow recombination hotspot for deletions of exons 2 and 3 to two regions, to the first with contiguously grouped AluY(1), FLAM_C, AluY(2) in intron 1, and to the second with a single copy of AluY(3) in intron 3.
Although Resta et al. consider the high density of Alu elements as an important factor, they pointed out that it does not seem sufficient to explain the high level of NAHR events since several genes with high Alu content such as thymidine kinase or beta-tubulin are not characterized by a high incidence of this type of damage. They mention that characteristic DNA motifs or additional features like high GC content may contribute to the generation of recurrent rearrangements of the STK11 gene. In our opinion, besides aforementioned features, DNA methylation level may be considered as another related factor. A few studies showed the association between methylation status of repeat-rich regions and the occurrence of recombination (cross over) events [27–29]. Those observations, which indicate that hypermethylation may inhibit homologous recombination, might explain the participation of specific Alus, despite the presence of closer situated and highly homologous elements.
Concentration of CNV in the distal part of the gene led us to the hypothesis of possible participation of intra-intronic rearrangements of STK11 intron 1 in conditioning PJS, in some cases with so far undetected genetic basis of the disease. This 11,213-bp long fragment is located in the STK11 recombination hotspot and characterized by the highest accumulation of interspersed repeats within the gene, reaching 43 % of its sequence (GC content 58 %). It cannot be excluded that those rearrangements (either deletions, duplications, or e.g. inversions) within the intron, might interfere in the correct splicing or expression of STK11 thereby conditioning PJS.
Deletion of exons 2–3 of the STK11 gene may prove to be the most recurrent large rearrangement causing PJS. Furthermore, its prevalence is not restricted to a particular geographical region, but concerns the worldwide population. In addition, the sequences present in its BPs may be involved in a significant percentage of the remaining gene CNVs. It is rather clear that there are multiple mechanisms of CNVs formation, which include both recombination and replication-based mechanisms. Thus, as reported, copy number changes are not randomly distributed, but multiple genomic features can affect the probability of their occurrence [10]. The data currently available unambiguously suggest participation of Alu elements from the two studied regions in all deletions of exons 2–3 so far described, and we have also shown their involvement in the other two mutations. Literature data indicate that approximately 30 % of PJS cases are caused by CNVs. According to our study, a third involve Alu elements of interest, thus we can assume that over 10 % of all PJS cases are related to the mutations concerning those two regions. This gives a new insight into the conditioning of this rare disease and allows to introduce improvements in genetic diagnostics of the patients.
References
Homan M, Dolenc Strazar Z, Orel R (2005) Peutz–Jeghers syndrome. A case report. Acta dermatovenerologica Alpina, Panonica, et Adriatica 14:26–29
Mehenni H, Resta N, Guanti G, Mota-Vieira L, Lerner A, Peyman M, Chong KA, Aissa L, Ince A, Cosme A et al (2007) Molecular and clinical characteristics in 46 families affected with Peutz–Jeghers syndrome. Dig Dis Sci 52:1924–1933
Marignani PA (2005) LKB1, the multitasking tumour suppressor kinase. J Clin Pathol 58:15–19
Riegert-Johnson DL, Westra W, Roberts M (2012) High cancer risk and increased mortality in patients with Peutz–Jeghers syndrome. Gut 61:322 author reply 322–323
Kaluzny A, Matuszewski M, Wojtylak S, Krajka K, Cichy W, Plawski A, Gintowt A, Lipska BS (2012) Organ-sparing surgery of the bilateral testicular large cell calcifying sertoli cell tumor in patient with atypical Peutz–Jeghers syndrome. Int Urol Nephrol 44:1045–1048
Kilic-Okman T, Yardim T, Gucer F, Altaner S, Yuce MA (2008) Breast cancer, ovarian gonadoblastoma and cervical cancer in a patient with Peutz–Jeghers syndrome. Arch Gynecol Obstet 278:75–77
Aretz S, Stienen D, Uhlhaas S, Loff S, Back W, Pagenstecher C, McLeod DR, Graham GE, Mangold E, Santer R et al (2005) High proportion of large genomic STK11 deletions in Peutz–Jeghers syndrome. Hum Mutat 26:513–519
Chow E, Meldrum CJ, Crooks R, Macrae F, Spigelman AD, Scott RJ (2006) An updated mutation spectrum in an Australian series of PJS patients provides further evidence for only one gene locus. Clin Genet 70:409–414
Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME et al (2006) Copy number variation: new insights in genome diversity. Genome Res 16:949–961
Hastings PJ, Lupski JR, Rosenberg SM, Ira G (2009) Mechanisms of change in gene copy number. Nat Rev Genet 10:551–564
Jurka J (2004) Evolutionary impact of human Alu repetitive elements. Curr Opin Genet Dev 14:603–608
De Rosa M, Galatola M, Quaglietta L, Miele E, De Palma G, Rossi GB, Staiano A, Izzo P (2010) Alu-mediated genomic deletion of the serine/threonine protein kinase 11 (STK11) gene in Peutz–Jeghers syndrome. Gastroenterology 138:2558–2560
Borun P, Bartkowiak A, Banasiewicz T, Nedoszytko B, Nowakowska D, Teisseyre M, Limon J, Lubinski J, Kubaszewski L, Walkowiak J et al (2013) High resolution melting analysis as a rapid and efficient method of screening for small mutations in the STK11 gene in patients with Peutz–Jeghers syndrome. BMC Med Genet 14:58
Orellana P, Lopez-Kostner F, Heine C, Suazo C, Pinto E, Church J, Carvallo P, Alvarez K (2013) Large deletions and splicing-site mutations in the STK11 gene in Peutz–Jeghers Chilean families. Clin Genet 83:365–369
Resta N, Giorda R, Bagnulo R, Beri S, Della Mina E, Stella A, Piglionica M, Susca FC, Guanti G, Zuffardi O et al (2010) Breakpoint determination of 15 large deletions in Peutz–Jeghers subjects. Hum Genet 128:373–382
Papp J, Kovacs ME, Solyom S, Kasler M, Borresen-Dale AL, Olah E (2010) High prevalence of germline STK11 mutations in Hungarian Peutz–Jeghers Syndrome patients. BMC Med Genet 11:169
Yang HR, Ko JS, Seo JK (2010) Germline mutation analysis of STK11 gene using direct sequencing and multiplex ligation-dependent probe amplification assay in Korean children with Peutz–Jeghers syndrome. Dig Dis Sci 55:3458–3465
Hearle NC, Rudd MF, Lim W, Murday V, Lim AG, Phillips RK, Lee PW, O’Donohue J, Morrison PJ, Norman A et al (2006) Exonic STK11 deletions are not a rare cause of Peutz–Jeghers syndrome. J Med Genet 43:e15
de Leng WW, Westerman AM, Weterman MA, Jansen M, van Dekken H, Giardiello FM, de Rooij FW, Paul Wilson JH, Offerhaus GJ, Keller JJ (2007) Nasal polyposis in Peutz–Jeghers syndrome: a distinct histopathological and molecular genetic entity. J Clin Pathol 60:392–396
de Leng WW, Jansen M, Carvalho R, Polak M, Musler AR, Milne AN, Keller JJ, Menko FH, de Rooij FW, Iacobuzio-Donahue CA et al (2007) Genetic defects underlying Peutz–Jeghers syndrome (PJS) and exclusion of the polarity-associated MARK/Par1 gene family as potential PJS candidates. Clin Genet 72:568–573
Volikos E, Robinson J, Aittomaki K, Mecklin JP, Jarvinen H, Westerman AM, de Rooji FW, Vogel T, Moeslein G, Launonen V et al (2006) LKB1 exonic and whole gene deletions are a common cause of Peutz–Jeghers syndrome. J Med Genet 43:e18
Jiang CY, Esufali S, Berk T, Gallinger S, Cohen Z, Tobi M, Redston M, Bapat B (1999) STK11/LKB1 germline mutations are not identified in most Peutz–Jeghers syndrome patients. Clin Genet 56:136–141
Borun P, Kubaszewski L, Banasiewicz T, Walkowiak J, Skrzypczak-Zielinska M, Kaczmarek-Rys M, Plawski A (2014) Comparativehigh resolution melting: a novel method of simultaneous screening for small mutations and copy number variations. Hum Genet 133:535–545
Bejjani BA, Shaffer LG (2006) Application of array-based comparative genomic hybridization to clinical diagnostics. J Mol Diagn JMD 8:528–533
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30:e57
D’Haene B, Vandesompele J, Hellemans J (2010) Accurate and objective copy number profiling using real-time quantitative PCR. Methods 50:262–270
Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J (2012) Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Nat Acad Sci USA 109:5880–5885
Maloisel L, Rossignol JL (1998) Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev 12:1381–1389
Colot V, Maloisel L, Rossignol JL (1999) DNA repeats and homologous recombination: a probable role for DNA methylation in genome stability of eukaryotic cells. J Soc Biol 193:29–34
Acknowledgments
This work was supported by the Ministry of Education and Science, Poland [Grant Number N402431438] and National Science Centre, Poland [Grant Number 2013/09/N/NZ5/02505]. The studies were approved by the local Ethics Committee of the Poznan University of Medical Science and performed after obtaining written informed consent from the patients.
Conflict of interest
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Borun, P., De Rosa, M., Nedoszytko, B. et al. Specific Alu elements involved in a significant percentage of copy number variations of the STK11 gene in patients with Peutz–Jeghers syndrome. Familial Cancer 14, 455–461 (2015). https://doi.org/10.1007/s10689-015-9800-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-015-9800-5