Abstract
Genetic resistance to the parasitic root-lesion nematode, Pratylenchus thornei, is one of the main management strategies cereal growers can use to minimise the impact of nematodes on winter cereal cropping. Screening of genotypes in the presence of P. thornei populations must provide reliable resistance measures that are realised under field conditions. Adoption of the latest statistical methodologies can help to better differentiate between resistant and susceptible genotypes. In this study, post-harvest P. thornei population densities were measured from a collection of 17 field experiments, with varying starting P. thornei population densities, conducted between 2011 and 2018 in locations across the northern grain growing region of eastern Australia. The experiments primarily consisted of wheat genotypes. The post-harvest P. thornei population densities were analysed across multiple environments in a linear mixed model framework, with a factor analytic structure used to model genotype by environment (G \(\times\) E) interaction effects exclusively for wheat genotypes. In general, genetic correlations between environments were found to be high, indicating limited G \(\times\) E interaction for resistance to P. thornei. Post-processing of results using the factor analytic selection tools (FAST) method provided a measure of the overall performance for each wheat genotype, as well as a stability measure reflecting the consistency of the resistance status across environments. The FAST method quantified genotype resistance on a continuous scale, better reflecting the nature of genetic resistance based on a quantitative variable such as nematode population density, and provided a statistically robust and informative means of aiding selection decisions for resistance to P. thornei.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Root-lesion nematodes (RLNs) are soilborne migratory endoparasites that penetrate plant roots and intercept the flow of water and nutrients into the plant, causing a subsequent negative effect on plant health and grain yield (Trudgill 1991). In favourable conditions, the ability of RLNs to reproduce more than once in a growing season allows population densities to rapidly increase (Reeves et al. 2020). The rate at which RLN population densities increase differs substantially between plant hosts, depending on their susceptibility or resistance to the RLN species (Jones and Fosu-Nyarko 2014; Owen et al. 2014; Thompson et al. 2008; Vanstone et al. 2008).
Root-lesion nematodes are found across the grain growing regions of Australia (Thompson et al. 2008; Vanstone et al. 2008). In the subtropical, northern grain growing region of eastern Australia (from latitudes of approximately -23.53°N to approximately -32.25°N), the most common species of RLN is Pratylenchus thornei (Thompson et al. 2010). The dominant winter cereal crops grown in the region include bread wheat (Triticum aestivum), durum wheat (Triticum durum) and barley (Hordeum vulgare), which are all known hosts of P. thornei.
Resistant genotypes of these crops are grown to control P. thornei population density increases (Fanning et al. 2018; Owen et al. 2014). Genotypes tolerant to P. thornei will maintain grain yield despite the presence of potentially damaging population densities of the nematode in the soil, but may still allow population densities to increase (Fanning et al. 2020; Trudgill 1991). Genetic diversity for both resistance and tolerance is evident within the winter cereal crops grown in the region. Resistant genotypes offer an effective management option for controlling nematode populations (Fanning et al. 2018; Owen et al. 2014; Reeves et al. 2020; Robinson et al. 2019), while tolerant genotypes ensure continued crop production in the presence of higher population densities.
There are multiple rigorous and repeatable methods of quantifying P. thornei population densities from soil samples. Since the late 1990s, molecular polymerase chain reaction (PCR) based methods have been developed to quantify a wide range of soilborne pathogen populations more accurately and quickly (Seesao et al. 2017). A method widely adopted in Australia is the PreDicta® B testing service, which uses quantitative PCR (qPCR) to estimate the total amount of P. thornei DNA detected in a soil sample via a DNA extraction system (Ophel-Keller et al. 2008). The service is used routinely in research experiments to quantify levels of a pathogen/s following the application of experimental treatments, and is particularly useful for resistance screening of genotypes for soilborne pathogens such as P. thornei (Fanning et al. 2018; Reeves et al. 2020). It is known that P. thornei resistance is fundamentally continuous in nature, due to the polygenic and subsequently quantitative nature of the trait (Trudgill 1991; Zwart et al. 2004). As such, the measurement using qPCR (e.g. PreDicta® B) of a continuous trait such as P. thornei population densities is well suited to the investigation of the continuum of P. thornei resistance.
In a research setting, resistance testing of genotypes to soilborne pathogens can be performed either in controlled environment (CE) experiments or in field experiments. In the case of RLNs, and in particular P. thornei, resistance testing under CE conditions is well developed (Sheedy et al. 2015; Thompson et al. 2020). One advantage of controlled conditions is that a consistent number of P. thornei can be added to the soil (Sheedy et al. 2015; Thompson et al. 1999), providing uniform P. thornei densities across all the experiment. However, it is also important to conduct resistance testing in the field, as the resistance status of genotypes must be realised under field conditions, where environmental factors may vary (Fanning et al. 2018). While field experiments provide natural growing conditions for both the crop and the nematode populations, the pathogen populations in the soil cannot be managed to be as uniform as in CE experiments.
Currently, the routine screening of Australian commercial and soon to be released bread wheat genotypes for resistance to P. thornei is primarily undertaken using CE experiments (Thompson et al. 2020). Nonetheless, multiple studies exploring the resistance of genotypes to P. thornei have also been performed using field-based experiments (Fanning et al. 2018; Owen et al. 2014), while others have explored the consistency of genetic resistance between CE and field-based experiments (Rodda et al. 2016; Thompson et al. 2020).
In the field, it is important to quantify genetic resistance under different environmental conditions through a series of experiments conducted across different locations and years, where the combined experiments form a multi-environment trial (MET) series. Statistical methods for the analysis of MET datasets are well-documented and are commonly formulated in a linear mixed model (LMM) framework (Smith et al. 2005). The LMM framework is powerful, as it enables the modelling of genotype by environment (G \(\times\) E) interaction effects while allowing for appropriate modelling of experimental design effects and spatial effects for each experiment (Cullis et al. 2010; Smith et al. 2001). The factor analytic (FA) variance structure has been widely used to model GxE interaction and has been shown to provide accurate MET analysis results for grain yield (Kelly et al. 2007), and more recently for RLN resistance (Fanning et al. 2018; Thompson et al. 2020).
The development of the factor analytic selection tools (FAST) post-processing methodology has improved the interpretability of the FA model outputs. The FAST method summarises the G \(\times\) E interaction pattern using the FA regression parameters to form measures of overall genotype performance and stability (Smith and Cullis 2018). To the best knowledge of the authors, there have been no previous reports of the FAST method being applied to MET datasets with the aim of determining genetic resistance to P. thornei, or RLNs in general; rather, this method is primarily designed for use within crop improvement programs (Cocks et al. 2019; Harris et al. 2019; Sjoberg et al. 2021). The FAST method has the potential to provide two simple metrics for the quantification and selection of genetic resistance to P. thornei, which retain important information regarding the continuous nature of overall performance and stability of genotypes.
In previous P. thornei CE experiments, it has been found that resistance rankings of bread wheat genotypes are quite stable across multiple experiments, due to a distinct lack of interaction in post-harvest P. thornei densities for genotypes across experiments (Sheedy et al. 2015; Thompson et al. 2011, 2020). In addition, minimal G \(\times\) E interaction in P. thornei population densities was previously reported from a set of field experiments conducted in southern Australia (Fanning et al. 2018). Similar field-based results from the testing of post-harvest P. thornei densities, across multiple field experiments in differing locations and years, has not yet been reported in the subtropical northern grain growing region of eastern Australia.
This study presents results from a MET analysis of post-harvest P. thornei population densities to quantify the genetic resistance of bread wheat genotypes to P. thornei in the subtropical northern grain growing region of eastern Australia. The aims of this study are to (i) explore genetic resistance to P. thornei of a set of bread wheat genotypes tested across multiple field experiments in the target environment, and (ii) demonstrate the novel application of the FAST post-processing methodology in the context of P. thornei resistance. The outputs from the FAST method aid genetic selection for resistance to P. thornei, providing metrics that respect the continuum of resistance. The analysis approach, coupled with the FAST post-processing methodology presented in this research, can potentially be applied to any RLN and/or crop species.
Materials and methods
Characterisation of experimental sites
The MET data comprised 17 field experiments conducted between 2011 and 2018, with experimental locations spread geographically from southern Queensland to central New South Wales. Characteristics of each experiment including the soil type (Webb et al. 1997), sowing and harvest dates, annual rainfall for the year of the experiment and mean yield are shown in Table 1. Also presented are the mean pre-sowing and post-harvest P. thornei population densities, which from this point on will be referred to as ‘initial’ and ‘final’ P. thornei population densities, respectively.
The original research aims of these experiments varied (Table 1). Eight experiments were conducted under the National Variety Trials (NVT) testing system (https://nvt.grdc.com.au/), which primarily aims to assess the yield potential of near to release and commercial genotypes of the major Australian grain crops in various agricultural regions across Australia. In all eight of these experiments, only bread wheat genotypes were tested and included in the MET analysis. A second subset of seven experiments aimed to test genetic variation in the tolerance of cereal genotypes to the soilborne disease crown rot, caused by the fungal pathogen Fusarium pseudograminearum. In each of these experiments, genotypes were exposed to a high level of crown rot pathogen burden, via inoculation with the pathogen at sowing (Dodman and Wildermuth 1987; Forknall et al. 2019), versus a control treatment with no applied inoculum. The aim in experiment NA13 was to test the impact of differential times of sowing (TOS) on genotype performance, with bread wheat and durum wheat genotypes sown at early, standard and late planting windows for the region in which the experiment was conducted. Another experiment, TU15, was designed to explore the impact of TOS on the genetic tolerance of cereal genotypes to crown rot, with genotypes exposed to a high level of crown rot pathogen burden, versus a control treatment, at two different planting times. A common characteristic of all experiments is the identification of significant initial P. thornei densities in bulk soil sampling prior to, or at sowing, as part of the site characterisation process, and the subsequent measurement of final P. thornei densities at the plot level, for assessing genetic resistance to P. thornei using qPCR (Table 1). The exception to this was experiment BE14, where P. thornei population density measurements were only conducted for the bread wheat genotypes and plots to which the control treatment (no applied crown rot inoculum) was allocated.
Experimental designs
Details of the experimental dimensions and parameters are given in Table 2. The NVT experiments included in the MET dataset were arranged according to randomised complete block designs, with blocking in two directions often implemented using the design package DiGGer (Coombes 2019). Experiment NA13 was arranged according to a split plot design, with the TOS treatments randomly allocated to main plots within replicate blocks, and the genotypes randomly allocated to sub plots within each main plot. The crown rot tolerance experiments (Table 1) were arranged according to randomised complete block designs, with the combinations of crown rot inoculum treatments and genotypes randomly allocated to plots within replicate blocks. The TOS by crown rot experiment, TU15, was arranged according to a split plot design, with the TOS treatments randomly allocated to main plots within replicate blocks, and the combinations of crown rot inoculum treatments and genotypes randomly allocated to sub plots within each main plot.
Experimental material
The genotypes tested were mostly bread wheats, with some additional barley and durum wheat genotypes included in approximately half of the experiments (Table 2). The investigation of G \(\times\) E interaction effects for the barley and durum crop types was not a priority in this MET analysis. This was because there were relatively low numbers of barley and durum genotypes tested in most experiments (with the exception of the 24 barley genotypes tested in TU15 (Table 2)), and there was low concurrence of genotypes belonging to these crop types between experiments. The bread wheat genotype concurrence between experiments is presented in Appendix Table 6. The number of unique bread wheat genotypes present in each experiment (at least 12), as well as the concurrence of bread wheat genotypes between experiments (at least four, except between MN15E and MN15M), enabled a robust assessment of bread wheat G \(\times\) E interaction effects for final P. thornei population densities.
Measurement of P. thornei populations
Initial P. thornei population densities were measured for each experiment as part of the site characterisation process. Between 20 and 30 soil cores (either 0–15 cm or 0–30 cm soil depth) were taken per replicate block, at sowing. Final P. thornei population densities were measured within zero to 60 days post-harvest, depending on adequate soil moisture to allow for collection of field samples. Between eight and 20 soil cores were taken from each plot (either 0–15 cm or 0–30 cm soil depth). The final soil samples were then combined within each plot to provide one sample per plot for testing. The PreDicta® B method was used to extract and quantify the concentration of P. thornei DNA in the soil samples using qPCR. This concentration was converted to a population density of P. thornei (P. thornei/g soil) using a standard conversion based on soil type (Ophel-Keller et al. 2008).
Statistical methods
The final P. thornei population densities per gram of soil were analysed in a LMM framework. A square root transformation of the raw data was necessary to satisfy the model assumptions of normality and homogeneity of variance across the range of fitted values. Heterogeneity of variance was over-corrected by a natural logarithm transformation, thus the square root transformation was favoured (Welham et al. 2014).
In this study, the term ‘environment’ was defined as the unique condition under which multiple genotypes were tested. The treatment levels for different times of sowing or different crown rot inoculum levels were concatenated with experimental location to define a specific environment, resulting in 28 unique environments (see EnvID in Table 2). The LMM for the MET data included fixed effects for crop type, experiment and their interaction. Design effects were included as random effects in the model, and spatial effects were modelled on an experiment basis (see ExpID in Table 2), following the methods of Gilmour et al. (1997). Heterogeneity of residual variance was accounted for at an experiment level. The random G \(\times\) E interaction effects were modelled on an environment basis, rather than on an experiment basis. Three variance structures were used to separately model the genetic effects for each of the three different crop types across environments. Random terms for both barley and durum genotype effects were fitted, as well as corresponding terms for the interactions between these and the environments that they were tested in, using a simple variance component model (Patterson et al. 1977).
An FA model was fitted to the G \(\times\) E interaction effects for bread wheat (Smith et al. 2001). The FA model consists of an underlying regression framework for the G \(\times\) E interaction effects based on a multiplicative combination of (environment) loadings by (genotype) scores. The factor loadings were rotated according to a varimax rotation (Smith et al. 2001). The percentage of repeatable G \(\times\) E variance accounted for by each factor (or by the multiplicative part of the FA model) is denoted as %VAF.
The genetic variance models for the barley and durum crop types were kept relatively simple due to the low number of genotypes of these crop types appearing in some experiments, and low genotype concurrence between environments. A variance structure with homogeneous genetic variance across environments and homogeneous covariance between environments was found to be most parsimonious for these crop types. All further reference to outputs from the fitting of FA models and mention of genetic or G \(\times\) E variation corresponds to that of the bread wheat crop type alone.
Factor analytic models of increasing order (denoted by FAk, where k corresponds to the order of model fitted) were iteratively fitted to the G \(\times\) E interaction effects. The most appropriate order of FA model was selected based upon the agreement of both the residual maximum likelihood ratio test (Stram and Lee 1994) and the Akaike Information Criterion (AIC) (Akaike 1973), with a decision based on the AIC favoured when the selection criteria indicated different models. Empirical best linear unbiased predictions (eBLUPs) were generated from the final model for the random G \(\times\) E interaction effects. Empirical best linear unbiased estimates (eBLUEs) were generated for the environment by crop type interaction (fixed) effects and variance components were estimated using residual maximum likelihood (Patterson and Thompson 1971). All models were fitted using the ASReml-R package (Butler et al. 2018), in the R computing environment (R Core Team 2020).
Outputs from the FA model can be complex and challenging to interpret without robust post-processing tools. The FAST method (Smith and Cullis 2018) allows for the regression implicit in the FA model to be summarised with two key results, namely the overall performance (OP) and the stability of each genotype in its responses to differing environments. The OP of a genotype is calculated as the mean of the rotated estimated loadings for the first factor, multiplied by the rotated score, also for the first factor, for that genotype. The interpretation of this result as the overall, or average, performance of a genotype is predicated on the fact that (after factor rotation) the first factor of the FA model explains the majority of the repeatable G \(\times\) E variation. If all the environmental loadings for the first factor are positive, then an assumption can be made that the first factor represents non-crossover G \(\times\) E interaction variation (Smith and Cullis 2018). Often, this coincides with a relatively high correlation between the first factor loadings and the mean of the response variable for each environment (Smith et al. 2015).
Following from this interpretation of OP, subsequent factors capture the remaining repeatable G \(\times\) E variation and are indicative of stability in the OP measure of a genotype. If a relatively large proportion of the repeatable G \(\times\) E variability for a genotype is accounted for by multiple factors, then its OP measure is likely to be less stable. The root mean square deviation (RMSD) of a genotype can be interpreted as its overall stability, with further details given in Smith and Cullis (2018). The FAST method therefore produces two meaningful measures, OP and RMSD, that can directly contribute to the quantification and selection of genotypes for resistance to P. thornei across multiple environments.
In the context of P. thornei resistance, the OP metric can be used to provide a continuum of genetic resistance, along which genotypes that comprised the MET dataset can be individually ranked. This OP continuum allows for a relative comparison of the resistances of all genotypes. Additionally, it allows for head-to-head pairwise comparisons of genotypes through testing the null hypothesis of equal OPs to determine statistically significant differences in P. thornei resistance (Welham et al. 2014). This head-to-head testing requires standard errors of the OPs (square root of the prediction error variance), which can be estimated using the predict.asreml function of ASReml-R (Butler et al. 2018).
The extent of G \(\times\) E interaction can be further investigated with a plot of the eBLUPs for a set of genotypes of interest at each of the environments (Smith et al. 2015). This allows visualisation of consistency of eBLUP rankings between environments. Environments can be arranged according to an agglomerative hierarchical clustering process (Kaufman and Rousseeuw 2009) based on the genetic correlations, to improve identification of crossover trends between clusters of environments. Clustering was performed in this study using the agnes package (Maechler et al. 2019) in the R statistical computing environment (R Core Team 2020).
Comparison of MET analysis results with industry reporting guidelines
In Australia, information on the resistance of genotypes to disease is delivered to industry using what are referred to as resistance ratings (https://nvt.grdc.com.au/nvt-disease-ratings). This resistance rating system consists of nine discrete ordinal categories, with genotypes given a rating ranging from what is labelled as “resistant” to “very susceptible”. In the case of resistance information related to P. thornei, ratings are defined by the equidistant division of the range of predicted final P. thornei population densities for the genotypes, based on predictions from a MET analysis (Thompson et al. 2020). Pratylenchus thornei resistance ratings for the genotypes that comprised the MET dataset are presented in Appendix Table 8, which have been collated from multiple sources, including the NVT website (https://nvt.grdc.com.au/nvt-disease-ratings), and regional crop variety sowing guides (Albatross Rural Consulting 2019, 2020; Lush 2016; Matthews and McCaffery 2019; Matthews et al. 2016, 2017, 2018, 2020, 2021), where data was retrieved on 13 January 2023. The results from the MET analysis were compared with the sourced resistance ratings for the genotypes that comprised the MET dataset.
Results
The iterative process of fitting FA variance models to the G \(\times\) E interaction effects is outlined in Table 3. An FA model of order 4 (FA4) was selected as the most parsimonious model, as it was optimal according to both the residual maximum likelihood ratio test and the AIC (Table 3).
A summary of the parameters from the model fitted to the MET dataset used to study the resistance of bread wheat genotypes to P. thornei is presented in Table 4. Three-quarters of the environments (21 out of 28) had greater genetic variance than residual variance. All environments had positive loadings for the first factor (Table 4), and all except three environments had 100% of their genetic variance explained by all four factors (Appendix Table 7). The overall %VAF by each factor was 79.39% for the first factor, 10.20% for the second factor, 7.33% for the third factor and 2.54% for the fourth factor, resulting in 99.46% of the G \(\times\) E interaction variation accounted for by the multiplicative, or repeatable, part of the FA model (Appendix Table 7). The correlation between the predicted final P. thornei densities for each environment and the first factor loadings (Table 4) was 0.85.
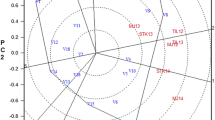
The genetic correlations between environments in the MET analysis are presented as a heatmap in Fig. 1. The environments included in the MET analysis were generally highly correlated in terms of final P. thornei population densities, with almost 80% of the pairwise genetic correlations between environments being greater than 0.5 (Fig. 1). The correlations ranged from a minimum of − 0.266 to a maximum of 0.991. Both environments in the WO18 experiment displayed weak negative correlations with two other environments (BU12 and NA13-TOS2) ranging from − 0.198 to − 0.266. There were no other negative correlations observed between any of the other pairs of environments.
Heatmap of genetic correlations between each pair of environments (labelled “EnvID”, see Table 2 for details), for the bread wheat genotypes tested in the analysis of the multi-environment trial dataset used to study the resistance of bread wheat genotypes to Pratylenchus thornei. Correlations range between − 1 and 1. A correlation of 1 between two environments indicates a perfect match of genotype rankings with respect to final P. thornei population densities; a correlation of − 1 indicates a complete reversal of genotype rankings, and a correlation of 0 indicates no relationship between the genotype rankings between environments
The positive loadings for all environments for the first factor (Table 4) of the FA4 model fitted to the G \(\times\) E interaction effects indicated that the first factor represented non-crossover G \(\times\) E interaction. This enabled a comparison of individual genotypes using the metrics of OP and RMSD (Fig. 2; data for Fig. 2 is supplied in Appendix Table 8).
Overall performance (OP) for each of the 118 bread wheat genotypes, plotted against their respective root mean square deviation (RMSD), from the factor analytic selection tools post-processing of the results from the analysis of the multi-environment trial dataset used to study the resistance of bread wheat genotypes to Pratylenchus thornei. The OP values are indicative of the overall resistance to P. thornei of a genotype. Large positive values indicate increased susceptibility to P. thornei, while large negative values indicate increased resistance, compared to the average performance of all genotypes tested in the dataset. The RMSD values indicate the stability of the predicted OP values; the higher the RMSD, the more unstable the resistance of a genotype. Genotypes of interest have been labelled with grey text and indicated with grey dots
The bread wheat genotype Strzelecki was the genotype with the largest positive OP (Fig. 2). This indicates that Strzelecki was the most susceptible genotype to P. thornei, resulting in the highest final P. thornei population density of the genotypes tested. Conversely, the genotype Suntop had the largest negative OP, meaning that it was the most resistant genotype in terms of final P. thornei population density in the set of bread wheat genotypes tested. Both Strzelecki and Suntop have moderately high RMSD values, with a higher RMSD than approximately 79 and 66% of genotypes tested, respectively. These high RMSD values indicate that Strzelecki and Suntop show moderate instability in their genetic resistance across environments. The genotype Mitch had the highest RMSD value of all the genotypes tested. Although its OP was higher than the average, its susceptibility was quite unstable, demonstrating sensitivity to varying environmental conditions.
The eBLUPs of final P. thornei population density for two selected subsets of genotypes are displayed for each environment (Fig. 3), where the two subsets contained genotypes with higher and lower stability of resistance, respectively. Genotypes in the two subsets displayed a wide range of predicted OPs, from − 0.93 to 0.99 for the stable genotypes, and from − 0.68 to 1.28 for the unstable genotypes. The figure provides further insight into the stability of performance of particular genotypes across particular environments, where there was greater cross-over interaction for the unstable genotypes between environments.
Empirical best linear unbiased predictions (eBLUPs) of the bread wheat genotype by environment (G \(\times\) E) interaction effects obtained from the analysis of post-harvest Pratylenchus thornei population density for two subsets of bread wheat genotypes, in each environment (EnvID) considered in the analysis of the multi-environment trial dataset investigating the resistance of bread wheat genotypes to P. thornei. The empirical best linear unbiased estimates (eBLUEs) of the post-harvest square root P. thornei population densities for each environment are given in parentheses following each EnvID label on the horizontal axis. Larger black dots indicate that a genotype was tested in that environment, while smaller grey dots indicate that the genotype was not tested in that environment. The two subsets of genotypes were selected to separate genotypes with higher and lower stability of resistances, to enable visualisation of this stability across environments. The eBLUPs of the G \(\times\) E interaction effects are presented as positive or negative deviations from the mean for each environment. The horizontal dashed line at zero denotes the overall mean for each environment. The environments are ordered along the horizontal axis according to an agglomerative hierarchical clustering algorithm (Kaufman and Rousseeuw 2009), such that environments that have more similar genotype rankings are clustered together
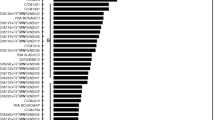
The current NVT resistance ratings (https://nvt.grdc.com.au/resources/disease-rating-definitions) are compared with the continuous OP resistance statuses obtained in this study (Fig. 4). The top facet of Fig. 4 shows the OPs of the commercial genotypes tested in this MET dataset, where the bars have been coloured according to the P. thornei resistance rating assigned by the NVT resistance rating system (Appendix Table 8). When genotypes are ordered in terms of their OP, there is only marginal correspondence between the OPs and the assigned NVT resistance ratings. The bottom facet of Fig. 4 shows a similar plot, where bars are instead coloured by the head-to-head comparisons of each genotype with a particular reference genotype, EGA Gregory. This genotype was chosen for comparison, as it was tested in the majority of environments and is also routinely used as a standard check for P. thornei resistance in the northern grain growing region of Australia by industry. These comparisons allow quick identification of genotypes that are predicted to be significantly more or less resistant to P. thornei than the widely grown genotype EGA Gregory, in terms of OP.
Graph of the overall performance (OP) values for each of the commercial bread wheat genotypes considered in the analysis of the multi-environment trial dataset used to study the resistance of bread wheat genotypes to Pratylenchus thornei. Error bars correspond to the standard error of the OPs. Genotypes have been ordered from lowest to highest OP, with a lower OP indicating greater resistance to P. thornei and a higher OP indicating greater susceptibility to P. thornei. In (a), the bars are coloured according to the P. thornei resistance ratings that have been assigned by the National Variety Trial (NVT) testing system (Appendix Table 8). Of the nine possible categorical resistance ratings assigned by NVT, only seven were observed in the set of genotypes considered. Resistance rating definitions can be found here https://nvt.grdc.com.au/resources/disease-rating-definitions. In (b), the bars are coloured according to whether or not they result in a significantly different OP compared to a reference genotype, EGA Gregory. The vertical red dashed line indicates the position of EGA Gregory on the horizontal axis
The key results of OP and RMSD can be condensed into a simple lookup table presenting the head-to-head comparisons of all commercial genotypes with three “check genotypes”, which span the range of resistance to P. thornei (Table 5). The genotypes used for comparison for this MET dataset were Suntop (resistant), EGA Gregory (average), and Strzelecki (susceptible). The results are coloured by RMSD to give an indication of stability of performance across environments. Table 5 is presented in a similar way to the current NVT sowing guides, however with resistance ratings replaced with head-to-head comparisons to a set of check genotypes. The information presented in Table 5 is subjective, and the authors suggest careful consideration of check genotypes, colour schemes, and other graphical tools when deciding how best to present the key information from the MET analysis.
Discussion
Cereal growers aiming to identify genotypes resistant to P. thornei require resistance information that is relevant under field conditions and is also straightforward to interpret. The application of the FAST method in this study has shown that it is an effective tool for quantifying genetic resistance to P. thornei in bread wheat genotypes through a MET analysis of final P. thornei population densities. The results provide a useful and practical summary of genetic resistance in terms of OP and RMSD, thereby providing cereal growers with the information they need to make informed bread wheat genotype selection decisions with respect to P. thornei resistance.
Comparison of the outputs of this study with both the literature, and communications to industry
Genetic resistance to P. thornei is informed by final P. thornei population densities, which are measured on a continuous scale. Thus, it is proposed that comparisons and selections should be made between genotypes on such a scale, rather than attempting to convert resistances to a discrete rating scale. In the recent literature on P. thornei resistance, analysis methods employed in studies such as Sheedy et al. (2015), Fanning et al. (2018) and Thompson et al. (2020) produced final P. thornei population densities predicted for each genotype at the average environment, on a continuous scale. Currently in Australia, through the NVT testing system, these predictions are then converted to a discrete rating scale based on nine equal subranges (equidistant divisions) of the range of predictions, respective to each testing region of Australia (Thompson et al. 2020). This conversion to discrete rating scales is consistent with other conventions of communicating genotype resistance information internationally (Agriculture and Horticulture Development Board 2021; Onofre et al. 2021). In creating the discrete rating labels, the actual range of final P. thornei population densities is obscured from interpretation. This might be taken to suggest that the discrete rating labels are relatable from one analysis to another, which is unlikely to be the case as both environmental conditions and the set of genotypes being tested are known to influence the range of final P. thornei population densities. Thus, the provision of resistance information on a continuous scale is vital to the interpretation of genetic resistance to P. thornei.
The FAST post-processing methodology was well suited in this study for summarising the important findings from the FA model on a continuous scale (Smith and Cullis 2018). Primarily, this was due to the relatively low level of crossover G \(\times\) E interaction, along with the fact that all the loadings on the first factor of the FA model were positive (Table 4), the %VAF by the first factor was substantial (79.39%), and additionally, there was a strong positive correlation (0.85) between the first factor loadings and the eBLUEs of the square root final P. thornei population densities for each environment. The use of the FAST approach was an improvement over the post-processing methods employed by Sheedy et al. (2015), Fanning et al. (2018) and Thompson et al. (2020). This is due to the appropriate separation of the factor that accounts for non-crossover G \(\times\) E interaction from the remaining factors, facilitating separate calculations of the metrics of overall genotype performance and stability (Smith and Cullis 2018). In this way, the metric of overall performance is not clouded by crossover G \(\times\) E interaction, however limited that crossover interaction may be, therefore more accurately capturing the genotypes’ “inherent resistance”. Additionally, it provided a widely applicable set of metrics obtainable through the FAST method (OP and RMSD) for informing P. thornei resistance.
Genotypes with high RMSD values (indicating higher variability in performance across environments) can be investigated further, to provide information on observed performance in individual environments. This is necessary to give context to the OP for these genotypes and allow growers to make more informed decisions regarding genotype resistance within particular environments. Graphical displays where the zero-centred eBLUPs of the G \(\times\) E interaction effects for each environment are plotted for selected genotypes (such as Fig. 3), provide an intuitive way to visualise this variability in performance (Smith et al. 2015). This visualisation provides a practical tool by which to gauge the G \(\times\) E interaction of interest, by involving only genotypes which are relevant to the decision being made. This approach is an improvement over both the use of overall genotype predictions averaged across environments, and the application of the discrete rating labels that are the current industry standard in Australia, due to their lack of information about variation in performance across environments.
In addition to overall inferences about P. thornei resistance, the method implemented in this study also facilitates specific genotype selections through head-to-head statistical comparisons of genotypes using their OPs. This is a key point of difference in the implementation of the FAST method in this study, compared to previous uses of the method where the main aim is genetic selection for subsequent progression within a crop improvement program. In the traditional setting for the FAST method, a plot such as that given in Fig. 2 provides all the necessary guidance to inform selection decisions. However, in this setting, where the aim is two-fold: (i) understanding the resistance of genotypes in the population, and (ii) performing head-to-head comparisons which determine statistical significance of a difference in resistance from one genotype to another, further exploration of the OPs is warranted using an approach such as that graphically illustrated by Fig. 4b.
This head-to-head comparison method is likely better suited to genotype selection in the context of commercial cropping, where selection for P. thornei resistance is likely not the only aim, and genotype selection may have already been narrowed to a handful of genotypes due to other selection criteria. The current industry standard of using discrete resistance rating labels for genotypes does not provide any measure of uncertainty or variability, thereby rendering head-to-head comparisons using these discrete labels potentially misleading (as shown in Fig. 4a). In contrast, the head-to-head comparison method detailed in this study retains transparency around the confidence in the determination of the overall resistance to P. thornei of any bread wheat genotype.
Using the head-to-head comparisons with check genotypes, a simple lookup table which condenses the key results from the FAST post-processing methodology can be constructed (Table 5). In this way, P. thornei resistance, and the stability of resistance across environments, can be communicated succinctly without having to rely on converting results to a discrete rating scale. Given that OP is a metric that is relative to the MET dataset under consideration, it is more appropriate to summarise resistance information by presenting key head-to-head comparisons with check genotypes that have been chosen to span the range of resistances observed in the MET dataset. This reduces the tendency for genotype resistance to be taken out of context of the MET analysis at hand. It also allows for uncertainty in cases where a particular genotype has not received sufficient testing; in that case, a genotype’s performance is more likely to be identified as not significantly different from that of the check genotypes. Additionally, the colour-coding of results based on RMSD indicates how reliable the result is expected to be across all environments. Lighter-coloured results suggest to the reader to be more cautious with their selection of that genotype, and perhaps investigate its resistance in particular environments further by using the information in Fig. 3, for example. It is envisaged that Table 5 could be included in sowing guides, as an alternative to current resistance ratings.
Crown rot and time of sowing environmental conditions
The repurposing of the experiments in the dataset for the investigation of P. thornei resistance provided a valuable opportunity to investigate G \(\times\) E interaction between different environmental conditions of interest within some of the field experiments, namely the different crown rot and time of sowing treatments (identified in Table 1). Most of the environments inoculated with crown rot recorded lower predicted mean final P. thornei population densities than their corresponding uninoculated environments (Table 4). Despite this, the ranking of bread wheat genotypes remained similar between crown rot treatments (with correlations from 0.79 to 0.99, Fig. 1). Conversely, the differences in predicted mean final P. thornei population densities between different TOS changed in nature between the two experiments where they were tested (Table 4), and the genetic correlations between different TOS were lower, although still moderate (0.61–0.77, Fig. 1). This suggests that TOS may affect both P. thornei population densities and genetic resistance to P. thornei more than the presence or absence of crown rot. A potential explanation for this is the longer growth time afforded to plants under the earlier TOS treatments, which would allow the P. thornei more time to infest the plant roots and multiply (Reeves et al. 2020) but also result in different environmental conditions during crop growth due to planting earlier in the season (Hunt et al. 2019).
Recommendation for the comparison of resistance testing between controlled environment and field settings
Quantitative PCR testing can be used to measure P. thornei population densities in both field and CE experiments (Fanning et al. 2018; Sheedy et al. 2015), making possible a comparison of genetic resistance statuses between both types of experiments. For a robust comparison to be made, data from all experiments (both field and CE based) should be jointly analysed in an LMM framework. This was done appropriately in chickpeas (Cicer arietinum) by Rodda et al. (2016), however their study only included one CE experiment, limiting the conclusions that could be made about genetic resistance more broadly. A more substantial balance of both field and CE experiments was presented in the bread wheat P. thornei resistance study by Thompson et al. (2020), however their analysis approach only incorporated CE experiments in the LMM framework. Their subsequent post-hoc approach to compare results from the CE experiments with the field experiments failed to account for different sources of variability in the field, and to estimate robust genetic correlations between the different environments. In order to effectively validate resistance statuses generated from CE experiments with field results (or vice versa), a suitable number of both types of experiments should be conducted and the data combined together in the one MET analysis using a LMM framework.
Conclusions
The MET analysis of final P. thornei population density measurements from the replicated field experiments in the dataset considered, along with the application of the FAST post-processing methodology, respects the continuous nature of genetic resistance to P. thornei in bread wheat. The high level of consistency in genotype performance observed across environments, in terms of P. thornei resistance, was evident from the relatively small proportion of crossover G \(\times\) E interaction detected, even between very different environmental conditions including TOS or background disease interactions. This limited amount of crossover GxE interaction lent itself to the application of the FAST post-processing methodology, enabling a purpose-built assessment of genotype performance which separates the genotypes’ inherent resistance from their potential interactions with different environmental conditions. In practice, the results from the application of this methodology will allow growers to assess the robustness of their cereal genotype selections in the context of both OP and stability of P. thornei resistance across environments, and to make head-to-head comparisons between genotypes on a continuum of P. thornei resistance status.
Data availability
Data will be made available on reasonable request.
Abbreviations
- AIC:
-
Akaike information criterion
- CE:
-
Controlled environment
- eBLUP:
-
Empirical best linear unbiased prediction
- eBLUE:
-
Empirical best linear unbiased estimate
- FA:
-
Factor analytic
- FAST:
-
Factor analytic selection tools
- G \(\times\) E:
-
Genotype by environment
- LMM:
-
Linear mixed model
- MET:
-
Multi-environment trial
- NVT:
-
National Variety Trials
- OP:
-
Overall performance
- qPCR:
-
Quantitative polymerase chain reaction
- RLN:
-
Root-lesion nematode
- RMSD:
-
Root mean square deviation
References
Agriculture and Horticulture Development Board (2021) AHDB recommended lists for cereals and oilseeds 2021/22: Summer edition. AHDB Cereals & Oilseeds, Kenilworth, Warwickshire, UK
Akaike H (1973) Maximum likelihood identification of Gaussian autoregressive moving average models. Biometrika 60(2):255–265. https://doi.org/10.1093/biomet/60.2.255
Albatross Rural Consulting (2019) 2019 Queensland winter crop variety sowing guide. Grains Research and Development Corporation
Albatross Rural Consulting (2020) 2020 Queensland winter crop sowing guide. Grains Research and Development Corporation
Butler DG, Cullis BR, Gilmour AR, Gogel BJ, Thompson R (2018) ASReml-R Reference Manual Version 4. Version 4 edn. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK
Cocks NA, March TJ, Biddulph TB, Smith AB, Cullis BR (2019) The provision of grower and breeder information on the frost susceptibility of wheat in Australia. J Agric Sci 157(5):382–398. https://doi.org/10.1017/S0021859619000704
Coombes N (2019) DiGGer: DiGGer design generator under correlation and blocking. R package version 1.0.4. edn
Cullis BR, Smith AB, Beeck CP, Cowling WA (2010) Analysis of yield and oil from a series of canola breeding trials. Part II. Exploring variety by environment interaction using factor analysis. Genome 53(11):1002–1016
Dodman R, Wildermuth G (1987) Inoculation methods for assessing resistance in wheat to crown rot caused by Fusarium graminearum group 1. Aust J Agric Res 38(3):473–486. https://doi.org/10.1071/AR9870473
Fanning J, Linsell K, McKay A, Gogel B, Munoz Santa I, Davey R, Hollaway G (2018) Resistance to the root lesion nematodes Pratylenchus thornei and P. neglectus in cereals: Improved assessments in the field. Appl Soil Ecol 132:146–154. https://doi.org/10.1016/j.apsoil.2018.08.023
Fanning JP, Reeves KL, Forknall CR, McKay AC, Hollaway GJ (2020) Pratylenchus thornei: the relationship between presowing nematode density and yield loss in wheat and barley. Phytopathology 110(3):674–683. https://doi.org/10.1094/phyto-08-19-0320-r
Forknall CR, Simpfendorfer S, Kelly AM (2019) Using yield response curves to measure variation in the tolerance and resistance of wheat cultivars to Fusarium crown rot. Phytopathology 109(6):932–941. https://doi.org/10.1094/phyto-09-18-0354-r
Gilmour AR, Cullis BR, Verbyla AP (1997) Accounting for natural and extraneous variation in the analysis of field experiments. J Agric Biol Environ Stat 2(3):269–293. https://doi.org/10.2307/1400446
Harris C, Boschma S, Brennan M, Borg L, Harden S, Cullis B (2019) Leucaena shows potential in Northern Inland New South Wales, Australia. Trop Grassl Forrajes Trop 7:120–126
Hunt JR, Lilley JM, Trevaskis B, Flohr BM, Peake A, Fletcher A, Zwart AB, Gobbett D, Kirkegaard JA (2019) Early sowing systems can boost Australian wheat yields despite recent climate change. Nat Clim Change 9(3):244–247. https://doi.org/10.1038/s41558-019-0417-9
Jones MGK, Fosu-Nyarko J (2014) Molecular biology of root lesion nematodes (Pratylenchus spp.) and their interaction with host plants. Ann Appl Biol 164(2):163–181. https://doi.org/10.1111/aab.12105
Kaufman L, Rousseeuw PJ (2009) Finding groups in data: an introduction to cluster analysis, vol 344. John Wiley & Sons, Hoboken
Kelly AM, Smith AB, Eccleston JA, Cullis BR (2007) The accuracy of varietal selection using factor analytic models for multi-environment plant breeding trials. Crop Sci 47(3):1063–1070. https://doi.org/10.2135/cropsci2006.08.0540
Lush D (2016) Queensland 2016 wheat varieties. Grains Research and Development Corporation and the Queensland Department of Agriculture and Fisheries (DAF)
Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K (2019) Cluster: cluster analysis basics and extensions. R package, Version 2.1.0. edn
Matthews P, McCaffery D (2019) Winter crop variety sowing guide 2019. New South Wales Department of Primary Industries, a part of New South Wales Department of Industry
Matthews P, McCaffery D, Jenkins L (2016) Winter crop variety sowing guide 2016. New South Wales Department of Primary Industries, as part of New South Wales Department of Industry, Skills and Regional Development
Matthews P, McCaffery D, Jenkins L (2017) Winter crop variety sowing guide 2017. New South Wales Department of Primary Industries, as part of New South Wales Department of Industry
Matthews P, McCaffery D, Jenkins L (2018) Winter crop variety sowing guide 2018. New South Wales Department of Primary Industries, a part of New South Wales Department of Industry
Matthews P, McCaffery D, Jenkins L (2020) Winter crop variety sowing guide 2020. New South Wales Department of Primary Industries, a part of the Department of Regional New South Wales
Matthews P, McCaffery D, Jenkins L (2021) Winter crop variety sowing guide 2021. New South Wales Department of Primary Industries, a part of the Department of Regional New South Wales
Onofre KA, De Wolf ED, Lollato R, Whitworth RJ (2021) Wheat variety disease and insect ratings 2021. Kansas State University
Ophel-Keller K, McKay A, Hartley D, Herdina CJ (2008) Development of a routine DNA-based testing service for soilborne diseases in Australia. Australas Plant Pathol 37(3):243–253. https://doi.org/10.1071/AP08029
Owen KJ, Clewett TG, Bell KL, Thompson JP (2014) Wheat biomass and yield increased when populations of the root-lesion nematode (Pratylenchus thornei) were reduced through sequential rotation of partially resistant winter and summer crops. Crop Pasture Sci 65(3):227–241. https://doi.org/10.1071/CP13295
Patterson HD, Silvey V, Talbot M, Weatherup STC (1977) Variability of yields of cereal varieties in UK trials. J Agric Sci 89(1):239–245. https://doi.org/10.1017/S002185960002743X
Patterson HD, Thompson R (1971) Recovery of inter-block information when block sizes are unequal. Biometrika 58(3):545–554. https://doi.org/10.1093/biomet/58.3.545
R Core Team (2020) R: a language and environment for statistical computing, 3.6.3 edn. R Foundation for Statistical Computing, Vienna, Austria
Reeves KL, Forknall CR, Kelly AM, Owen KJ, Fanning J, Hollaway GJ, Loughman R (2020) A novel approach to the design and analysis of field experiments to study variation in the tolerance and resistance of cultivars to root lesion nematodes (Pratylenchus spp.). Phytopathology 110(10):1623–1631. https://doi.org/10.1094/phyto-03-20-0077-r
Robinson NA, Sheedy JG, Macdonald BJ, Owen KJ, Thompson JP (2019) Tolerance of wheat cultivars to root-lesion nematode (Pratylenchus thornei) assessed by normalised difference vegetation index is predictive of grain yield. Ann Appl Biol 174(3):388–401. https://doi.org/10.1111/aab.12504
Rodda MS, Hobson KB, Forknall CR, Daniel RP, Fanning JP, Pounsett DD, Simpfendorfer S, Moore KJ, Owen KJ, Sheedy JG, Thompson JP, Hollaway GJ, Slater AT (2016) Highly heritable resistance to root-lesion nematode (Pratylenchus thornei) in Australian chickpea germplasm observed using an optimised glasshouse method and multi-environment trial analysis. Australas Plant Pathol 45(3):309–319. https://doi.org/10.1007/s13313-016-0409-4
Seesao Y, Gay M, Merlin S, Viscogliosi E, Aliouat-Denis CM, Audebert C (2017) A review of methods for nematode identification. J Microbiol Methods 138:37–49. https://doi.org/10.1016/j.mimet.2016.05.030
Sheedy JG, McKay AC, Lewis J, Vanstone VA, Fletcher S, Kelly A, Thompson JP (2015) Cereal cultivars can be ranked consistently for resistance to root-lesion nematodes (Pratylenchus thornei & P. neglectus) using diverse procedures. Australas Plant Pathol 44(2):175–182. https://doi.org/10.1007/s13313-014-0333-4
Sjoberg SM, Carter AH, Steber CM, Garland Campbell KA (2021) Application of the factor analytic model to assess wheat falling number performance and stability in multienvironment trials. Crop Sci 61(1):372–382. https://doi.org/10.1002/csc2.20293
Smith A, Cullis B, Thompson R (2001) Analyzing variety by environment data using multiplicative mixed models and adjustments for spatial field trend. Biometrics 57(4):1138–1147. https://doi.org/10.1111/j.0006-341X.2001.01138.x
Smith AB, Cullis BR (2018) Plant breeding selection tools built on factor analytic mixed models for multi-environment trial data. Euphytica 214(8):143. https://doi.org/10.1007/s10681-018-2220-5
Smith AB, Cullis BR, Thompson R (2005) The analysis of crop cultivar breeding and evaluation trials: an overview of current mixed model approaches. J Agric Sci 143(6):449–462. https://doi.org/10.1017/S0021859605005587
Smith AB, Ganesalingam A, Kuchel H, Cullis BR (2015) Factor analytic mixed models for the provision of grower information from national crop variety testing programs. Theor Appl Genet 128(1):55–72. https://doi.org/10.1007/s00122-014-2412-x
Stram DO, Lee JW (1994) Variance components testing in the longitudinal mixed effects model. Biometrics 50(4):1171–1177. https://doi.org/10.2307/2533455
Thompson JP, Brennan PS, Clewett TG, Sheedy JG, Seymour NP (1999) Progress in breeding wheat for tolerance and resistance to root-lesion nematode (Pratylenchus thornei). Australas Plant Pathol 28(1):45–52. https://doi.org/10.1071/AP99006
Thompson JP, Clewett TG, Sheedy JG, Reen RA, O’Reilly MM, Bell KL (2010) Occurrence of root-lesion nematodes (Pratylenchus thornei and P. neglectus) and stunt nematode (Merlinius brevidens) in the northern grain region of Australia. Australas Plant Pathol 39(3):254–264. https://doi.org/10.1071/AP09094
Thompson JP, Owen KJ, Stirling GR, Bell MJ (2008) Root-lesion nematodes (Pratylenchus thornei and P. neglectus): a review of recent progress in managing a significant pest of grain crops in northern Australia. Australas Plant Pathol 37(3):235–242. https://doi.org/10.1071/AP08021
Thompson JP, Reen RA, Clewett TG, Sheedy JG, Kelly AM, Gogel BJ, Knights EJ (2011) Hybridisation of Australian chickpea cultivars with wild Cicer spp. increases resistance to root-lesion nematodes (Pratylenchus thornei and P. neglectus). Australas Plant Pathol 40(6):601. https://doi.org/10.1007/s13313-011-0089-z
Thompson JP, Sheedy JG, Robinson NA (2020) Resistance of wheat genotypes to root-lesion nematode (Pratylenchus thornei) can be used to predict final nematode population densities, crop greenness, and grain yield in the field. Phytopathology 110(2):505–516. https://doi.org/10.1094/phyto-06-19-0203-r
Trudgill D (1991) Resistance to and tolerance of plant parasitic nematodes in plants. Annu Rev Phytopathol 29:167
Vanstone VA, Hollaway GJ, Stirling GR (2008) Managing nematode pests in the southern and western regions of the Australian cereal industry: continuing progress in a challenging environment. Australas Plant Pathol 37(3):220–234. https://doi.org/10.1071/AP08020
Webb A, Grundy M, Powell B, Littleboy M (1997) The Australian sub-tropical cereal belt: soils, climate and agriculture. In: Sustainable crop production in the sub-tropics: an Australian perspective, pp 8–23
Welham SJ, Gezan SA, Clark SJ, Mead A (2014) Statistical methods in biology: design and analysis of experiments and regression. CRC Press, Boca Raton
Zwart RS, Thompson JP, Godwin ID (2004) Genetic analysis of resistance to root-lesion nematode (Pratylenchus thornei) in wheat. Plant Breed 123(3):209–212. https://doi.org/10.1111/j.1439-0523.2004.00986.x
Acknowledgements
The authors would like to acknowledge the Grains Research and Development Corporation (GRDC) for their investment in statistical research in agriculture through the project DAQ00208. The authors would like to acknowledge the New South Wales Department of Primary Industries and Northern Grower Alliance technical staff who helped to conduct the experiments included in the dataset. The authors would like to acknowledge Professor Richard Jarrett of the University of Adelaide, who provided extensive help with the formation of the manuscript through his writing workshop. The authors would also like to acknowledge very helpful feedback for the manuscript received from Gabriela Borgognone and Michael Mumford, as well as the members of the Leslie Research Facility Action Learning Group, including: Mandy Christopher, Jack Christopher, John Thompson, Andrew Zull and Kerry Bell. Additionally, the authors acknowledge helpful feedback from the journal review process, which considerably improved the quality of the paper.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by the Grains Research and Development Corporation (project codes DAV00128, DAN00175 and DAQ00208).
Author information
Authors and Affiliations
Contributions
AK and CF conceptualised the study. BR, CF and AK developed the statistical models, that BR, CF and LN then implemented to analyse the data. SS and RD conducted the field trials and collected the data for analysis. The first draft of the manuscript was written by BR and all authors commented on subsequent versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
See Appendix Tables 6, 7, and 8.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rognoni, B., Forknall, C.R., Simpfendorfer, S. et al. Quantifying the resistance of Australian wheat genotypes to Pratylenchus thornei based on a continuous metric from a factor analytic linear mixed model. Euphytica 220, 141 (2024). https://doi.org/10.1007/s10681-024-03387-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-024-03387-2