Abstract
Broomcorn millet (Panicum miliaceum L.) is a prehistorical cereal, today cultivated as a minor crop with low yields but with a renewed interest for its high water use efficiency and gluten-free grains. To reverse the downward trend in broomcorn millet cultivation, the crop needs genetic improvement and creation of novel genetic variation to increase productivity. In order to facilitate genomics-assisted breeding, we designed a reduced representation genome-sequencing assay that investigates 1.8% of the nuclear DNA in a targeted and reproducible way, with an intensity of genomic sampling that is a direct function of local recombination rate. We used this tool and set up bioinformatics analyses tailored to the polyploid genome of P. miliaceum for maternity and paternity testing, quantification and genomic distribution of homozygous regions and estimation of parental genome contribution for individual seedlings in advanced inbred lines from a breeding program and compared their genomic composition with registered varieties. We found several clues that suggest that the genetic purification process to ensure genetic uniformity is incomplete in varieties of this species. Residual heterozygosity was detected in the genome of three registered varieties ranging from 4.4 to 6.25% of their haploid genome length. Other registered varieties show genome-wide homozygosity. We found, however, evidence of intravarietal genetic variation in three cases that suggest that the breeder seed or commercial seed production had fixed by self-pollination multiple inbred lines with very similar, though not identical, genotypes within each variety.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Landraces and varieties of the species Panicum miliaceum L. are cultivated as a small-grain crop for human consumption and birdseed under the common name of broomcorn, proso or common millet, hereafter referred to as broomcorn millet. Despite having declined over time to a minor cereal, broomcorn millet is one of the world’s oldest domesticated plants (Lu et al. 2009) that provided food for prehistorical Eurasian populations (Hunt et al. 2011). Broomcorn millet is one of the most water use efficient grain crops and it is gluten free. Unlike other grains in which those traits are disjoint, broomcorn millet can provide solutions to two issues that agriculture and human nutrition will be confronted with in the near future: water scarcity in expanding semi-arid regions (Feng and Fu 2013; Yao et al. 2020) and immunological reaction in more people suffering from celiac disease (Lebwohl and Rubio-Tapia 2021). Part of the exceptionally low water requirement is due to the short growing cycle. Broomcorn millet produces harvestable grain within 60–90 days of sowing and currently serves either as a primary summer crop or as a catch and complement crop in the agricultural systems of China and North America.
P. miliaceum is a tetraploid species (2n = 4x = 36) that has originated from an allopolyploidization event, (Shi et al. 2019; Zou et al. 2019), combining rather recently (~ 0.5–2 million years ago) two genomes that had diverged for ~ 4.8–6.3 million years from their common ancestor (Sun et al. 2023; Wang et al. 2023). The P. miliaceum genome is composed by two sets of nine highly collinear homoeologous chromosomes, totaling approximately 900 Megabases (Mb) in size, donated by the species Panicum capillare and Panicum repens or their close relatives (Hunt et al. 2014). Two whole-genome assemblies were available in P. miliaceum at the time of this manuscript submission. One genome was sequenced from a landrace originating from Northern China (Zou et al. 2019). The other genome was sequenced from a Chinese cultivated variety (Shi et al. 2019). Both genomes were sequenced with PacBio long-reads that were assembled into chromosome-scale pseudomolecules showing comparable quality assembly metrics. We referenced the analysis of this paper to the GCA_003046395.2 Pm_0390_v2 assembly, corresponding to the NCBI BioProject PRJNA431363 and described in the paper of Zou et al. (2019). An improved version of the other genome (Shi et al. 2019; Wang et al. 2023) as well as novel genomes (Sun et al. 2023) were assembled more recently. Polyploids at early stages of diploidization, in which two or more sets of homoeologous chromosomes are originated from closely related species and homoeologous gene loss is still underway, pose the challenge of mapping uniquely and specifically DNA short-read alignments to either homoeolog in reference genome-based analyses. P. miliaceum showed a substantial level of sequence diversity between homoeologous chromosomes that should facilitate homoeologous-specific read mapping. However, incompleteness in the assembly and post-polyploidization deletions that are not fixed in the population, including those that may have occurred in the lineage of the reference individual, are technical and biological issues that generate homoeologous SNP calls, which determine what is referred to in other papers as “fixed heterozygosity” (Vischi et al. 2021), inflating nucleotide diversity estimates.
An accurate estimation of the level of nucleotide diversity in the cultivated germplasm is indeed missing, which may provide insight into the evolutionary events that have generated the present–day diversity and may guide breeding. The wild ancestor of the cultivated broomcorn millet is supposed to be P. miliaceum subsp. ruderale. The present-day populations of this subspecies in the presumed area of distribution, which may span a vast region in Eurasia from the Caspian Sea basin to China, are elusive. The unavailability of genuine wild individuals, descending from a common ancestor and surviving until the present in isolation from the crop germplasm, complicates the reconstruction of the history of domestication, improvement and cultivation of broomcorn millet (Stevens et al. 2021). Spontaneous broomcorn millet is commonly found in Eurasia and in North America (Bough et al. 2011) and is distinguished from cultivated germplasm by shattering panicles and low seed germination rates (Carpenter and Hopen 1985), which are typically wild traits. Genetically, spontaneous populations may consist of either genuine undomesticated individuals or feral and naturalized forms phenotypically indistinguishable from their wild counterparts, and mixtures of both categories. Only one recent report has suggested the occurrence of isolated populations of undomesticated P. miliaceum in Northern China that share very limited genetic ancestry with weedy and cultivated relatives in the rest of China and elsewhere (Xu et al. 2019). Questions remain to be answered as to whether low diversity is intrinsic and irremediable in this species (i.e. due to a single polyploidization event at the origin of the species) or declines in population size following domestication (Lu et al. 2009) and genetic drift during the westward expansion of the crop are reversible. It is, indeed, assumed that, as the crop was introduced into Central, Western and South Asia, European Russia and Eastern Europe, and from there into the North American high plains, genetic diversity was progressively lost.
Genetic diversity in broomcorn millet has been recently analyzed by genotyping–by–sequencing using three large but only partially overlapping germplasm collections. One is held at the Institute of Crop Science, Chinese Academy of Agricultural Sciences (ICSCAAS) in China (Li et al. 2021). Another one is held the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) in India (Johnson et al. 2019). The last one is held at the United States Department of Agriculture (USDA) in the US (Boukail et al. 2021). The ICSCAAS genebank, which totals 106 partially sequenced accessions, is highly enriched for weedy and cultivated forms from the center of domestication of the species in Eastern Asia. The ICRISAT genebank, which has 190 partially sequenced accessions, is enriched for cultivars grown in Western Asia and South Asia. The USDA genebank was represented in the sequenced set by 88 accessions mostly consisting of elite cultivars, including those selected by US breeders in the past 40 years. Other large collections of P. miliaceum, not included in any genomic surveys, are held at the Vavilov All—Russian Scientific Research Institute of Plant Industry in the Russian Federation and at two institutes in Ukraine (Goron and Raizada 2015). Due to the different genotyping-by-sequencing protocols, i.e. GBS (Elshire et al. 2011) with PstI, dRAD (Peterson et al. 2012) with HindIII and SLAF (Sun et al. 2013) with RsaI and and HaeIII restriction enzymes, used by the three research groups involved in these studies (Johnson et al. 2019; Li et al. 2021; Boukail et al. 2021), the SNP panels were also only partially overlapping. Single primer enrichment technology (SPET) sequencing, which we decided to use in this paper, is an alternative approach for reduced representation library sequencing that targets specific regions and sites in a highly reproducible way and with ample flexibility in terms of genome sampling size and depth of coverage (Scaglione et al. 2019).
Autogamous self-pollination is expected alone to ensure that landraces and varieties in P. miliaceum, which are propagated by seeding, are true-breeding lines. Molecular analyses with microsatellite markers seem to confirm homozygosity at all tested loci in a set of elite varieties and landraces (Flajšman et al. 2019; Vischi et al. 2021), though these assays investigate only an infinitesimal fraction of the genome, with only one conflicting report challenging these expectations and data (Xu et al. 2019). Intentional breeding based on controlled hybridization of divergent inbred lines has been conducted systematically in China, India, the US and Russia since only the past century (Gomashe 2017; Santra et al. 2019), leading to the release of registered varieties. For that purpose, distinctness, uniformity and stability (DUS) testing is mandatory and should guarantee that registered varieties breed true. It is common practice in millet hybridization breeding to perform 5–6 cycles of self-pollination and selection to generate quasi-inbred homozygous lines that are released as varieties. Breeders repeat selfing and selection until the seedlings show phenotypic uniformity and stability. DNA testing is not required to prove genome-wide homozygosity. However, residual heterozygosity in released varieties may later disclose segregation of phenotypic traits upon cultivation under previously untested circumstances, which is highly undesirable for both breeders and farmers.
With the dual-pronged aim of accelerating the genomics-assisted selection of purebreds and certifying the distinctness and homozygosity of P. miliaceum varieties prior to release, here we use SPET sequencing for assessing the breadth of preexisting diversity captured by an enterprise-run breeding program and the novel genetic variation generated through hybridization of divergent inbred lines. The analytical pipeline includes maternity and paternity testing, quantification and genomic distribution of homozygous regions and estimation of parental genome contribution for individual seedlings across generations. As a by-product of this genomic survey, we present several lines of evidence that suggest the need of handling with care seed propagation in germplasm repositories, release of new varieties and commercial seed production. We, indeed, found that the genetic composition of seed lots for recognized varieties may conceal, in some cases, the concomitant presence of highly similar although not exactly identical inbred lines or, in other cases, residual heterozygosity.
Materials and methods
Plant material and DNA sequencing
Seed lots of P. miliaceum varieties were obtained in the frame of the Re-Cereal Project funded by the European Fund for Regional Development (Interreg V-A Italia-Austria 2014–2020) as described in Vischi et al. (2021). Additional varieties were obtained from the seed companies Kriesel Certified Seed, Gurley, NE (Plateau) and Kutnowska Hodowla Buraka Cukrowego (KHBC) Sp. z o.o. Kłodawa, Poland (Gierczyckie) and from the food company MartinoRossi SpA, Malagnino, Italy (Dakota). F1 hybrids were generated by artificial hybridization following the procedure described in Nelson (1984). The subsequent generations of selfing were produced using a single-seed-descent scheme. Details of the sequenced accessions are reported in Supplementary Table S1. Seeds were germinated and seedlings were grown in 2 L potted soil (peat moss-based mix supplemented with compost, perlite, and starter fertilizer) maintained at soil water capacity in a greenhouse within a temperature range of 20–30 °C until sampling. From 50 to 70 days-old individual plants were used as a source material for DNA extraction, without bulking prior to or after DNA extraction, for both registered varieties and seedlings of the F1, F3, F4, F5, S1, S3 and S5 families. DNA was extracted from foliar tissues using the Sigma-Aldrige® GenElute Plant Genomic DNA Miniprep Kit (Merck KGaA, Darmstadt, Germany) kit. SPET oligonucleotides were synthesized by Tecan Trading AG (Männedorf, Switzerland). DNA libraries were generated using the Allegro Targeted Genotyping kit (Tecan Trading AG) with 70 ng of DNA as an input and following the manufacturer’s instructions. Paired-end reads of 150 bp in length each were obtained on an Illumina NovaSeq6000 sequencer. Library preparation and sequencing were outsourced to IGA Technology Services (Udine, Italy).
SPET design
In order to capture diversity in the gene space, while maintaining high conservation in primer annealing sites, we selected the longest intron in each gene and designed the probe in the adjacent 5′-flanking exon (Supplementary Fig. S1). In case of monoexonic genes, the probe was designed in the 5′-end of the exon with a 3′ to 5′ orientation in order to sequence the 5’–UTR. We also designed primers to interrogate 1.8 k variant sites that were polymorphic in ICSCAAS accessions, 1.8 k variant sites polymorphic in ICRISAT accessions and 2.2 k variant sites polymorphic in USDA accessions (Supplementary Fig. S2) in order to compare the accessions and breeding lines of this study with the germplasm included in genomic surveys as of September 2021 (Supplementary Methods).
Bioinformatics analyses
Raw sequences were trimmed for quality and adapter sequences using ERNE (Del Fabbro et al. 2013) and Cutadapt (Martin 2011) with default parameters. Primer sequences were not removed at this stage in order to use this information later for homoeologous–specific reads selection. Reads were aligned with the reference genome (GCA_003046395.2_Pm_0390_v2_genomic.fna) using BWA-MEM (Li and Durbin 2009) with default parameters. Only uniquely mapping reads (i.e. reads with a mapping quality > 10) were retained for further analyses. Raw variant sites were called using GATK HaplotypeCaller (Poplin et al. 2017). Individual genotypic calls were retained if read coverage was comprised between 10 and 1000 and quality parameters were QD < 2.0, MQ < 40.0, MQRankSum <-12.5. We used the thresholds of reference/alternative allele coverage ratio between 0.25 and 0.75 for calling heterozygous genotypes, ≥ 0.9 for calling homozygous reference genotypes and ≤ 0.1 for calling homozygous alternative genotypes. Variant sites were retained in the dataset if genotype calls passed the filters in > 50% of the accessions. Chromosomal coordinates refer to the GCA_003046395.2 reference genome assembly (Zou et al. 2019). Transposable elements were predicted using EDTA (Ou et al. 2019). Genomic windows-based analyses were conducted using a segmentation of the genome into 823 non-overlapping windows of variable size, containing 500 Kb of low-copy DNA each (Supplementary Fig. S3).
Filtering of false variant sites from aspecific read alignments between homoeologous copies
In order to remove homoeologous SNPs from SPET sequencing we performed a step of read alignment selection. We retained only first-in-pair reads (R1) that aligned at the expected primer start site (± 1 nt), considering the strand of the primer, and the corresponding second–in-pair reads (R2) originating from the same DNA fragment. Residual homoeologous variant sites called by GATK HaplotypeCaller were then removed from the vcf file using the information from selfed progeny of inbred lines (Sunup USDA, Quartet). In detail, we filtered out false polymorphic sites with a threshold of 5% of heterozygous genotypes calls in selfed progeny of inbred lines. We also used complete and validated pedigrees (P1, P2, F1, Fn) for removing polymorphic sites showing Mendelian inconsistency in at least one descendant. False variant sites residing in primer annealing sites and low quality variant sites falling within TE intervals, as identified by the software EDTA, were removed from the vcf file.
Comparison with genomic surveys that used other methods of genomic sampling
We detected 5179, 18,534 and 107,696 variants sites out of 1,943,371, 5,003,109 and 16,860,570 informative bases, respectively, in shared targeted regions between our SPET assay and the reduced representation genomic libraries obtained using GBS, dRAD and SLAF assays. In the pairwise comparison of specimens carrying the same genotype, which were subject to sequencing using two different assays (i.e. the varieties ‘Huntsman’ and ‘Sunrise’ that were sequenced using SPET and dRAD), we estimated a genotypic mismatch rate of one variant site out of 354,401 informative sites between replicates in ‘Huntsman’ and one variant site out of 362,859 informative sites between replicates in ‘Sunrise’.
Parentage testing
SNP genotypes were coded: homozygous reference equal to 2, homozygous alternative equal to 0 and heterozygous equal to 1. We considered only sites that are fully informative in each trio. An F1 hybrid is regarded as having passed the parental test if identity-by-state (IBS) was equal to zero at less than 1% of the informative sites in the trio among the sites that are polymorphic in the population and the IBS = 0 sites were randomly distributed across the genome. An F3, F4 and F5 individual is regarded as having passed the parental test if > 99% of the heterozygous sites showed IBS = 1 with both putative parents and > 99% of the homozygous sites in the descendant that showed IBS = 0 with either parent showed IBS = 2 with the other parent.
Parental genome contribution in advanced inbred lines
Parental state at each variant site in each trio was determined upon the following conditions. If (i) the genotype of the descendant is homozygous and identical to P1 and (ii) P1 is different from P2, then the parental state of the descendant is P1. If (i) the genotype of the descendant is homozygous and identical to P2 and (ii) P2 is different from P1, then the parental state is P2. If (i) the genotype of the descendant is heterozygous and (ii) P1 and P2 are homozygous for alternative alleles, then the parental state is hybrid. If (i) the genotype of the descendant is homozygous and P1 and P2 are homozygous and identical, then the parental state is identical to the parents. The parental genome composition was estimated based on the aggregated length of genomic intervals spanning consecutive sites showing the same parental state under the assumption of maximum parsimony in recombination events and was expressed as a percentage of the aggregated length of informative intervals.
Results
Targeted sequencing
We designed a panel of 45,004 SPET primers that target 39,247 nuclear genes for sequencing the gene space in P. miliaceum, using the genome assembly and the gene annotation of the BioProject PRJNA431363, and 5757 known variant sites in the P. miliaceum germplasm (Supplementary Table S2). With the sequencing output per sample obtained in this experiment (Supplementary Table S1), the SPET panel investigates a portion of the P. miliaceum genome that amounts to an average of 11.6 Mb, with a coverage comprised between 10 and 1000 reads in at least 50% of the samples, corresponding to 1.4% of the nuclear DNA. Approximately another 4 Mb (representing 0.4% of the nuclear DNA) were sequenced at the required read depth within these regions but they overlapped with predicted transposable elements and were not used for SNP calling. Supplementary Fig. S4 shows the distribution in size of the stacked read intervals in 40,492 informative genomic regions that were retained for analyses.
In the non-repetitive fraction of the genome, we identified 10,221 SNPs in our panel of 93 DNA samples, which included 12 unique genotypes and 4 redundant profiles, and 77 individuals in the line of descent of 4 putative parents. The first two components of a principal component analysis (PCA) explained cumulatively 27.3% of the variance and the PCA bidimensional plot in Fig. 1A was in agreement with most of the expected relations among accessions (Table 1). Another 2084 private SNPs were identified in the individual from which the reference genome had been assembled. Of all of these, 1736 SNPs were located in coding sequences, 5,400 SNPs in intronic regions, 1088 in UTRs and 4081 SNPs in the intergenic space. Nucleotide diversity in registered varieties was 2.3 × 10–4 in coding sequences, 1.9 × 10–4 in intronic regions, 2.6 × 10–4 in UTRs and 4.4 × 10–4 in the intergenic space. Considering the relative representation of informative bases in these four genomic contexts captured by our SPET sequencing compared to their relative abundance in the whole genome, we estimate an average nucleotide diversity of 3.93 × 10–4 with the genomic distribution shown in Supplementary Fig. S5. In the accession of P. miliaceum from which the genome reference sequence had been assembled (BioSample SAMN08389585), we detected a modal value of 94% sequence identity between homoeologous chromosomes (with the distribution shown in Supplementary Fig. S6) across the genomic intervals that were informative in SPET sequencing.
Principal component analysis (PCA) of the genetic diversity (A) and proportion of heterozygous variant sites in 93 P. miliaceum accessions (B). Solid circles in A and salmon background in B indicate existing varieties. The names of parental lines are bold faced in A. Colour in A indicates the family and the parental combination from which the progeny was derived. Asterisks in A mark samples that were renamed based on their SNP profiles, more details are provided in Table 1. Other symbols in A and background colour in B indicate generations. In B, yellow background indicates true-to-type F1 hybrids. Sea green background indicates F3 inbred lines. Olive green background indicates F4 inbred lines. Forest green background F5 inbred lines. The hash marks three released varieties with an above-the-background fraction of heterozygous sites. Asterisks and blue background highlight expected F1 hybrids and/or subsequent generations that turned out to be identical to the maternal inbred line
SNP variation
Based on breeder’s notes and suppliers’ seed dispatch records, we were expecting to be dealing with F3, F4 and F5 generations from 13 families originating through a single-seed descent (SSD) scheme from seven crosses, including one reciprocal cross, and 11 US, European and Russian varieties, including the putative parents of all crosses. An individual plant from the F1 generation was also included in the sequenced set for 7 out of 13 families. Raw counts of homozygous and heterozygous sites in each accession confirmed, in general, the composition of the germplasm under study (Supplementary Fig. S7). Registered varieties showed the lowest proportions of heterozygous sites, as expected for inbred lines (Fig. 1B). F1 hybrids showed the highest proportions of heterozygous sites, compatible with their origin from the crossing of distinct inbred lines. F3, F4, F5, lines showed intermediate proportions of heterozygous sites between parental lines and F1 hybrids, representing quasi-inbred lines. Exceptions to these rules emerged in every category. Three registered varieties stood out for a proportion of heterozygous sites above the background level as if they were quasi-inbred lines. One putative F1 hybrid and three SSD families showed a background level of heterozygous sites, as if they were derived from self-pollination of inbred lines. The most advanced inbred lines of F5 generations resulting from cross-mating between European and American varieties showed similar levels of heterozygosity as quasi-inbred lines that are already registered as varieties, though not yet as low as the background levels that are detected in most of the registered varieties.
Genome-wide homozygosity in registered varieties and advanced inbred lines
The varieties ‘Dawn’, ‘Horizon’, ‘Huntsman’, ‘Kornberger’, ‘Lisa’, ’Quartet’, ‘Sunrise’ and ‘Sunup’ showed a random genomic distribution of heterozygous SNPs, as expected in the case of false heterozygous genotypic calls in pure inbred lines (Supplementary Fig. S8). Two regions were included in this graphical representation and in the computation shown in Fig. 1B from the subtelomeric end of the upper arms of chromosome 4 (1.5 Mb) and chromosome 17 (575 Kb) that carried fixed heterozygosity in several varieties, including those that appear to be homozygous elsewhere in the genome as well as in segregating progenies. Fixed heterozygosity may have resulted in our analysis from either a homoeologous replacement, which is frequently found in other allopolyploids in subtelomeric regions (Scalabrin et al. 2020) as a result of homoeologous pairing and homoeologous recombination, or a large duplication. The varieties ‘Gierczyckie’, ‘Konstantinovsky’ and ‘Plateau’ are exceptions to genome-wide homozygosity because they showed 10, 14, and 16 chromosome segments, respectively, carrying a high frequency of heterozygous SNPs (Supplementary Fig. S8) that were arranged in a pattern that was compatible, in each variety, with the combination of two haplotypes present in the population. The varieties ‘Gierczyckie’, ‘Konstantinovsky’ and ‘Plateau’ are therefore not pure breeds. The seedlings of these varieties that we analyzed here still carry 38, 37.8 and 53.4 Mb of heterozygous DNA, respectively, corresponding to 4.4%, 4.4%, and 6.25%, respectively, of the haploid genome length, and should be considered advanced inbred lines in which the process leading to genetic purity is still incomplete. This is a precursor condition in recently selected varieties that may later develop into the situation that we observed in elder varieties, in which we assume that different haplotypes were alternatively fixed during seed production in a mixture of coexisting inbred lines collectively used by seed companies and farmers as archetypal for each variety (see below). Pairwise comparisons of purebreed varieties showed variable levels of homozygous differences (Fig. 2).
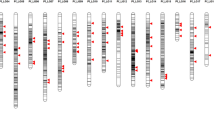
Homozygous variant sites in pairwise comparisons between varieties. A Pairwise comparisons between varieties used as parental lines in this study. B Other representative pairwise comparisons. Homozygous sites are plotted as red ticks in the right-hand sector of the diagram of each chromosome. The white-to-green heatmap in the left-hand sector of the diagram of each chromosome illustrates the number of informative sites in non-overlapping windows containing 500 Mb of low-copy DNA each
Maternity and paternity testing
Three out of 13 families and an F1 hybrid turned out to be S1, S3 and S5 generations that originated from ‘Quartet’ and ‘Sunup’ self-pollination. The genomic composition of the individuals within these families was revealed by their whole-genome homozygosity (Fig. 1B and Supplementary Figs. S9–S10A), the absence of homozygous differences in pairwise comparisons between siblings (Supplementary Figs. S10B–S11) and their maternal parent (Supplementary Fig. S11) and haplotype sharing with F3 descendants (Supplementary Fig. S10C). This was also confirmed by the absence of phenotypic segregation at a visual inspection of the adult plants (data not shown). We used all individuals of these families as genomic controls to remove false heterozygous variants sites that do not segregate and to determine intravarietal variation. We could confirm a hybrid origin for the remaining families.
We could confirm the expected parentage for nine families of hybrid origin. Of these, four families of hybrid origin showed consistent genotypic profiles with the plants used in this study as representative of the parental varieties. The remaining five families have derived from crosses in which either the maternal or the paternal line was not identical but showed limited differences with respect to the representative specimen sequenced in this study. We detected genetic variation (i.e. different haplotypes always present in a homozygous condition in each individual) among individual plants of the parental lines ‘Quartet’, ‘Sunup’ and ‘Lisa’ that were either sequenced here or whose genotypic profiles were inferred from their selfed progenies and/or from the offspring they generated by controlled hybridization.
Intravarietal variation
Two different inbred lines were found in ‘Quartet’. One inbred line is represented by the plant sequenced in this study (referenced to as Q1 in Table 1) and by the seed parent that originated the F1 hybrid “Q × L #C7” and one of the F3 families (family #127 in Table 1). The other ‘Quartet’ inbred line (referenced to as Q2 in Table 1) is represented by the seed parent that originated by self–pollination the invariant S1 and S3 lines sequenced in this study (Q × S #C10-4 and family #124 in Table 1). The inbred lines Q1 and Q2 differ for 855 variant sites out of 11,039,145 informative sites (Fig. 3). These variants sites showed Mendelian segregation and were not randomly distributed across the genome but they were located in at least 30 intervals of variable size, amounting to 230 Mb in aggregated size. There are at least two regions on the lower arms of chr9 and chr11, spanning respectively 2 Mb and 13 Mb and including collectively 58 variant sites, which showed patterns of SNPs compatible with two haplotypes being alternatively fixed in the two ‘Quartet’ inbred lines. For instance, across the concerned 2–Mb region on chr9, one ‘Quartet’ inbred line was homozygous for a haplotype that is shared with ‘Lisa’, ‘Gierczyckie’, ‘Horizon’ and ‘Sunup’, the other ‘Quartet’ inbred line was homozygous for a different haplotype shared with Dawn’, ‘Huntsman’, ‘Konstantinovsky’, ‘Kornberger’, ‘Sunrise’ and ‘Plateau’. Q2 and not Q1 was the inbred line that served as a pollen parent in the generation of other F1 hybrids. We, indeed, found evidence that the variant sites between ‘Lisa’ and Q2, but not between ‘Lisa’ and Q1, segregated only in the #120 F3 family and were invariant in ‘Lisa’, Q1, Q × L #C7 and all F3 individuals of the #127 family. Vice versa, the variant sites between ‘Lisa’ and Q1, but not between Lisa and Q2, were heterozygous in Q × L #C7 and segregated in the #127 family but not in the #120 family. The intervals carrying alternative haplotypes were small in terms of size and they were located in subtelomeric, gene-rich, TE-poor regions, which are supposed to correspond to highly recombinogenic euchromatic regions (Supplementary Fig. S12). The largest intervals carrying alternative haplotypes (i.e. on chromosomes 4 and 16) tend to span large centromeric and pericentromeric regions, which appeared to be gene–poor and rich either in tandem repeats or in TEs and are expected to correspond to low recombinogenic heterochromatic regions (Supplementary Fig. S12).
Homozygous variant sites in pairwise comparisons between different inbred lines within each variety. Homozygous sites are plotted as red ticks in the right-hand sector of the diagram of each chromosome. The white-to-green heatmap in the left-hand sector of the diagram of each chromosome illustrates the number of informative sites in non-overlapping windows containing 500 Mb of low-copy DNA each
Three different inbred lines coexist in ‘Sunup’. One inbred line is represented by the S3 and S5 families #111 and #119 (Table 1) that were obtained from the unintended self–pollination of ‘Sunup’. While all being fully homozygous and identical to one another, they did not match the genotypic profile of the ‘Sunup’ accession included in our set. The scale of genotypic mismatches (1806 variant sites out of 12,237,013 informative sites) and their genomic distribution (Fig. 3) suggest a similar situation as that observed for ‘Quartet’ and compatible with the hypothesis that they are similar inbred lines mixed up in the breeder seed of ‘Sunup’. We compared the invariant genotypic profiles of the families #111 and #119 with the genotypic profiles from other germplasm collections. They were identical with the ‘Sunup’ accession introduced from USDA and included in the set analyzed by Boukail et al. (2021) at all but four homozygous sites out of the 358,364 informative sites that were in common between our SPET experiment and dRAD sequencing (Supplementary Fig. S13B). All other F1 hybrids and advanced inbred lines in the present study that were originated from ‘Sunup’ show genotypic profiles that are compatible with being derived from a genotype identical to the families #111 and #119 and therefore identical to the ‘Sunup’ USDA accession (Table 1). The ‘Sunup’ accession in our set did not match any accession in the set of varieties analyzed by Boukail et al. 2021 which also includes varieties of similar origin (Supplementary Fig. S13A), reinforcing the hypothesis that they may represent coexisting inbred lines within the seed pool that is grown under the name ‘Sunup’ rather than a distinct misclassified variety. At a closer inspection, not even the donors of the ‘Sunup’ genome to the progenies of this study were plants raised from an absolutely uniform seed batch. From the genotypes in their progeny, we inferred the existence of two ‘Sunup’ inbred lines that differed for one variant site at the position chr15:17,287,776 out of 12,272,560 informative sites (Table 1, Fig. 3). This variation, although little, was confirmed by the evidence of Mendelian inheritance. One inbred line, which we refer to as Sunup USDA_b in Table 1, carries a GG genotype at the variant site and is the ancestor of all #119 S1 individuals, the Kornberger × Sunup F1 hybrid and the Lisa × Sunup F1 hybrid plant #C8-4. The same GG genotype is also found in all the F3, F4, F5 individuals of the #125 and #128 families that are homozygous for the Sunup haplotype. The other inbred line, which we refer to as Sunup USDA_a in Table 1, carries a CC genotype and is the ancestor of all #111 S1 individuals, the Lisa × Sunup F1 hybrid plant #C13-2 (genotype CG) and all the individuals of the #129 family that are homozygous for the Sunup haplotype.
Two different inbred lines also exist in ‘Lisa’. Three F1 hybrids and six families of advanced inbred lines descending from ‘Lisa’ perfectly matched across the parental genome contribution the specimen used in our set as a reference for ‘Lisa’, hereafter and in Table 1 referred to as L1. Two other F1 hybrids and two families of advanced inbred lines, which were expected to descend from crosses in which ‘Lisa’ served as a pollen parent, failed to pass the paternity test with L1 and were originated from what we consider another inbred line within the variety ‘Lisa’. Although we did not have in our panel another inbred line that matched this second ‘Lisa’ inbred line (which we referred to as L2), we inferred its genotype from their offspring for which the maternal parent ‘Kornberger’ was known and validated (i.e. K × L #C1-1 and K × L #C1-2 in Table 1). Based on this prediction, L1 and L2 are expected to differ for 512 variant sites out of 11,060,821 informative sites (Fig. 3).
Two plants that we obtained from two different commercial seed batches of ‘Kornberger’ differed for only 5 variant sites out of 12,062,306 informative sites. All the ‘Kornberger’ progeny we had in our set were derived from one of the genotypes. Unlike in ‘Sunup’ and ‘Lisa’, it was therefore impossible to validate those 5 variants sites as cases of intravarietal diversity and rule out the possibility that they may represent genotypic errors. Two plants that we obtained from two different commercial seed batches of ‘Huntsman’ were identical at all 11,755,274 informative sites. Two plants that we obtained from two different commercial seed batches of ‘Dawn’ were identical at all 11,755,274 informative sites. We also analyzed one plant that was raised from the germination of ‘Dakota’ millet (Fig. 2B), a commercial birdseed that is advertised on the packaging under this commercial name but it is believed to be obtained from the variety ‘Huntsman’, and found only 2 variants sites out of 11,760,619 and 12,053,362 informative sites, respectively, in comparison with our ‘Huntsman’ references.
Genomics-assisted selection of purebreeds from F1 hybrids
The genetic variation captured by the parental lines of our breeding program is comparatively ample if examined in relation to the P. miliaceum diversity collected in US, Chinese and Indian genebanks. While there is only little genetic overlap between our germplasm and Eastern and Southern Asian germplasm (Supplementary Figs. S14‒S15), the diversity that was generated by crossing our selected parental lines bridged the gap of genetic diversity between the currently used varieties in Western Eurasia and America (Supplementary Fig. S16). The expected heterozygosity in F1 hybrids was highest in crosses between Lisa and Sunup and lowest in crosses between Kornberger and Sunup (Fig. 1B). We validated the pattern of genomic distribution of alternatively homozygous sites predicted in the parental genomes with the observed distribution of heterozygous sites in the F1 offspring for three crosses (Supplementary Fig. S17). We then used SPET sequencing to monitor the effects of consecutive generations of self-pollination on the extent of runs of homozygosity in the progeny (Supplementary Table S3, Fig. 4A, Supplementary Figs. S18‒S25). The tabular and graphical outputs, which express and represent, respectively, the residual heterozygosity in terms of genome length are tools to be used at each round of selection to retain the lines that are closest to the condition of pure-breeding strains.
Residual heterozygosity (A) and parental genome composition (B) in F3 advanced inbred lines deriving from the cross between ‘Kornberger’ and ‘Quartet’. Genome-wide distribution of heterozygous sites (plotted as blue ticks in the right-hand sector of the diagram of each chromosome in A). Genome-wide distribution of maternal homozygous sites (plotted as pink ticks), paternal homozygous sites (plotted as cyan ticks) and heterozygous sites (plotted as yellow ticks in the right-hand sector of the diagram of each chromosome in B). Red ticks represent homozygous alternative sites between the parental inbred lines. The white-to-green heatmap in the left-hand sector of the diagram of each chromosome illustrates the number of informative sites in non-overlapping windows containing 500 Mb of low-copy DNA each in A and B
Variance of the parental genome contribution to inbred lines derived from biparental crosses
In the F3, F4 and F5 generations, we identified individuals with large variation in each parental genome contribution (i.e. those sampled from the tails of the normal distribution). With this analysis (Fig. 4B, Supplementary Figs. S26‒S29, Supplementary Tables S3‒S4), it is feasible to select from a single cross divergent inbred lines based on preferential maternal or paternal genome retention (i.e. to establish a backcross-like genetic background). This analysis will also offer the additional option to select for homozygosity inherited from either parent at target genetic intervals spanning specific genes, once beneficial allelic variants controlling trait variation will be known in this species (Rajput et al. 2016; Liu et al. 2022). In our advanced generation selfed lines, we observed large portions of intact parental chromosomes across all pericentromeric regions (exemplified by the case shown in Supplementary Fig. S30A‒B), which suggests that the generation of mosaic chromosomes that recombine fragments from divergent inbred lines is very difficult over these regions because of their intrinsically low recombination rate (Supplementary Fig. S30C).
Discussion
Our data show that the currently used varieties in broomcorn millet are, in general, pure inbred lines, therefore producing uniform progenies upon sexual propagation in the commercial activity of seed production. We found, however, residual heterozygosity in the varieties ‘Gierczyckie’ and ‘Konstantinovsky’ that were selected in Poland and Russia, respectively, as well as in ‘Plateau’, a waxy (amylose-free starch) grain variety registered in the US in 2015. This is not surprising for ‘Plateau’, which is an accession originally selected from advanced inbred lines of an F4 generation, following the intentional cross between ‘Huntsman’ and a Chinese waxy parent (Santra et al. 2015). Residual heterozygosity in advanced inbred progenies is commonplace in autogamous species such as soybean (Haun et al. 2011) and even in inbred lines of allogamous crops subject to intensive genomics–assisted breeding such as maize (Liu et al. 2018).
We detected a nearly perfect match for a number of accessions of the same variety (i.e. ‘Huntsman’, ‘Dawn’, ‘Sunrise’) within our set and in comparison with other studies, which excludes that technical flaws in the comparison of genotypic calls within and between different sequencing technologies have generated biases. We found some inconsistences in SNP genotype profiles between our set and published sequencing data (which we reanalyzed from raw sequencing reads in order to exclude technical artifacts due to different analytical pipelines, Boukail et al. 2021) that could be suggestive of curation errors in genebanks and in shipping of seed packets or inaccurate handling of seedlings in nursery beds. However, we observed that the non-matching SNP genotype profiles between samples taken from seedlings of one purportedly single variety are in most cases much more similar than non–matching SNP genotype profiles between samples taken from seedlings of different varieties. We are therefore inclined to believe that, in some instances, one variety is propagated as a mixture of genetically closely related inbred lines that are phenotypically indistinguishable and form a seemingly uniform pool of individuals, which satisfies DUS requirements. We found direct (comparison of sequenced individuals) or indirect (inferred parental genotypes from the progeny they generated) evidence of multiple inbred genotypes in plants raised from seed batches of the varieties ‘Quartet’, ‘Lisa’, ‘Sunup’.
Using this genomic survey in a breeding program, we monitored the effects of consecutive generations of self-pollination on the residual heterozygosity in advanced breeding lines originating from the parental combination of purebreds of different varieties with variable levels of homozygous variation. In addition to help selecting individuals with the most favorable genomic composition at each cycle of selection and although the following is not required by DUS testing protocols, these data strongly recommend the adoption of DNA testing at the end of the breeding process—before entering the stage of commercial seed production—to ensure that the breeder seed is a purebred. Residual heterozygosity that segregates during seed propagation and/or multiple similar inbred lines that coexist in a seed batch are not obstacles to varietal registration and certified seed production, provided that all of this does not translate into visible phenotypic differences. However, hidden genetic heterogeneity may result into phenotypic heterogeneity when a new variety is exposed to untested growing conditions in different environments or is confronted with unprecedented situations.
Our results also attest the effect of at least two technical and biological issues on slowing down P. miliaceum hybridization breeding. First, even following a well-established and carefully operated procedure of flower emasculation, complete prevention of self-pollination proved hard to be achieved in a massive breeding operation aimed at generating large controlled progenies. This observation may at least in part explain the limited progress in selecting higher yielding and more adapted progeny, despite the scale of past efforts, if breeders were in many cases selecting from families that largely derived from the unintended selfing of nearly homozygous parental lines. Second, despite hybridization between divergent parental lines is obtained, the nearly complete suppression of recombination across dozens of Mb tends to conserve intact parental chromosomes over multiple generations. This observation illustrates the difficulty in reshuffling genomes and creating mosaic patterns of chromosomal segments of different origins in P. miliaceum, in particular across the presumed pericentromeric regions. The extent and distribution of this phenomenon are indeed consistent with the reported intrachromosomal variation in recombination rate (Zou et al. 2019) and the affected regions correspond to large tracts of gene-poor and transposable element-rich heterochromatic DNA identified in the most complete and recent version of the P. miliaceum assembly (Wang et al. 2023). They are therefore predictable by computer simulations at the stage of breeding design.
In conclusion, the data we presented here underscores the persistent limits that are slowing down the progress of breeding and the genetic gain in a minor cereal species, which can be breached by resorting to genomic analyses that assist germplasm exploitation, progeny selection and certified seed production.
Data availability
DNA sequences are deposited in Sequence Read Archive (SRA) under the BioProject number PRJNA896096.
References
Bough M, Colosi JC, Cavers PB (2011) The major weedy biotypes of proso millet (Panicum miliaceum) in Canada. Can J Bot 64:1188–1198. https://doi.org/10.1139/B86-161
Boukail S, Macharia M, Miculan M et al (2021) Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.). BMC Plant Biol 21:1–12. https://doi.org/10.1186/s12870-021-03111-5
Carpenter J, Hopen H (1985) A comparison of the biology of wild and cultivated proso millet (Panicum miliaceum). Weed Sci 33:795–799
Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM (2013) An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS ONE 8:e85024. https://doi.org/10.1371/journal.pone.0085024
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. https://doi.org/10.1371/JOURNAL.PONE.0019379
Feng S, Fu Q (2013) Expansion of global drylands under a warming climate. Atmos Chem Phys 13:10081–10094. https://doi.org/10.5194/ACP-13-10081-2013
Flajšman M, Štajner N, Kocjan Ačko D (2019) Genetic diversity and agronomic performance of Slovenian landraces of proso millet (Panicum miliaceum L.). Turk J Bot 43:185–195
Gomashe SS (2017) Proso millet, Panicum miliaceum (L.): genetic improvement and research needs. In: Patil V (ed) Millets and sorghum: biology and genetic improvement. Wiley, New Jersey, pp 150–169
Goron TL, Raizada MN (2015) Genetic diversity and genomic resources available for the small millet crops to accelerate a new green revolution. Front Plant Sci. https://doi.org/10.3389/FPLS.2015.00157
Haun WJ, Hyten DL, Xu WW et al (2011) The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol 155:645. https://doi.org/10.1104/PP.110.166736
Hunt HV, Badakshi F, Romanova O et al (2014) Reticulate evolution in Panicum (Poaceae): the origin of tetraploid broomcorn millet, P. Miliaceum. J Exp Bot 65:3165. https://doi.org/10.1093/JXB/ERU161
Hunt HV, Campana MG, Lawes MC et al (2011) Genetic diversity and phylogeography of broomcorn millet (Panicum miliaceum L.) across Eurasia. Mol Ecol 20:4756. https://doi.org/10.1111/J.1365-294X.2011.05318.X
Johnson M, Deshpande S, Vetriventhan M et al (2019) Genome-wide population structure analyses of three minor millets: kodo millet, little millet, and proso millet. Plant Genome 12:190021. https://doi.org/10.3835/PLANTGENOME2019.03.0021
Lebwohl B, Rubio-Tapia A (2021) Epidemiology, presentation, and diagnosis of celiac disease. Gastroenterology 160:63–75. https://doi.org/10.1053/J.GASTRO.2020.06.098
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li C, Liu M, Sun F et al (2021) Genetic divergence and population structure in weedy and cultivated broomcorn millets (Panicum miliaceum L.) revealed by specific-locus amplified fragment sequencing (SLAF-Seq). Front Plant Sci 12:1242. https://doi.org/10.3389/fpls.2021.688444
Liu N, Liu J, Li W et al (2018) Intraspecific variation of residual heterozygosity and its utility for quantitative genetic studies in maize. BMC Plant Biol 18:1–15. https://doi.org/10.1186/s12870-018-1287-4
Liu T, Liu X, He J et al (2022) Identification and fine-mapping of a major QTL (PH1.1) conferring plant height in broomcorn millet (Panicum miliaceum). Front Plant Sci. https://doi.org/10.3389/FPLS.2022.1010057
Lu H, Zhang J, Liu KB et al (2009) Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc Natl Acad Sci U S A 106:7367–7372. https://doi.org/10.1073/pnas.0900158106
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Nelson LA (1984) Technique for crossing proso millet. Crop Sci 24:205–206. https://doi.org/10.2135/CROPSCI1984.0011183X002400010049X
Ou S, Su W, Liao Y et al (2019) Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol 20:1–18. https://doi.org/10.1186/s13059-019-1905-y
Peterson BK, Weber JN, Kay EH et al (2012) Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135. https://doi.org/10.1371/journal.pone.0037135
Poplin R, Ruano-Rubio V, DePristo MA et al (2017) Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. https://doi.org/10.1101/201178
Rajput SG, Santra DK (2016) Schnable J (2016) Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). Mol Breed 364(36):1–18. https://doi.org/10.1007/S11032-016-0460-4
Santra DK, Heyduck RF, Baltensperger DD et al (2015) Registration of “Plateau” waxy (amylose-free) proso millet. J Plant Regist 9:41–43. https://doi.org/10.3198/jpr2013.11.0067crc
Santra DK, Khound R, Das S (2019) Proso millet (L.) breeding: progress, challenges and opportunities. Adv Plant Breed Strateg Cereal 5:223–257. https://doi.org/10.1007/978-3-030-23108-8_6
Scaglione D, Pinosio S, Marroni F et al (2019) Single primer enrichment technology as a tool for massive genotyping: a benchmark on black poplar and maize. Ann Bot 124:543–552. https://doi.org/10.1093/aob/mcz054
Scalabrin S, Toniutti L, Di Gaspero G et al (2020) A single polyploidization event at the origin of the tetraploid genome of Coffea arabica is responsible for the extremely low genetic variation in wild and cultivated germplasm. Sci Rep 101(10):1–13. https://doi.org/10.1038/s41598-020-61216-7
Shi J, Ma X, Zhang J et al (2019) Chromosome conformation capture resolved near complete genome assembly of broomcorn millet. Nat Commun 101(10):1–9. https://doi.org/10.1038/s41467-018-07876-6
Stevens CJ, Shelach-Lavi G, Zhang H et al (2021) A model for the domestication of Panicum miliaceum (common, proso or broomcorn millet) in China. Veg Hist Archaeobotan 30:21–33. https://doi.org/10.1007/s00334-020-00804-z
Sun Y, Liu Y, Shi J et al (2023) Biased mutations and gene losses underlying diploidization of the tetraploid broomcorn millet genome. Plant J 113:787–801. https://doi.org/10.1111/TPJ.16085
Sun X, Liu D, Zhang X et al (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8:e58700. https://doi.org/10.1371/JOURNAL.PONE.0058700
Vischi M, Zorzin N, Bernhart M et al (2021) Morphophysiological and molecular characterization of millet (Panicum miliaceum L.) varieties for crop improvement in Western Europe. CABI Agric Biosci 21(2):1–12. https://doi.org/10.1186/S43170-021-00047-6
Wang Z, Huang S, Yang Z et al (2023) A high-quality, phased genome assembly of broomcorn millet reveals the features of its subgenome evolution and 3D chromatin organization. Plant Commun. https://doi.org/10.1016/J.XPLC.2023.100557
Xu Y, Liu M, Li C et al (2019) Domestication and spread of broomcorn millet (Panicum miliaceum L.) revealed by phylogeography of cultivated and weedy populations. Agronomy 9:835. https://doi.org/10.3390/AGRONOMY9120835
Yao J, Liu H, Huang J et al (2020) Accelerated dryland expansion regulates future variability in dryland gross primary production. Nat Commun 111(11):1–10. https://doi.org/10.1038/s41467-020-15515-2
Zou C, Li L, Miki D et al (2019) The genome of broomcorn millet. Nat Commun 101(10):1–11. https://doi.org/10.1038/s41467-019-08409-5
Acknowledgements
We thank Società Agricola Odorico and Maurizio Martinuzzi for plant nurturing and IGA Technology Services for DNA sequencing.
Funding
Open access funding provided by Università degli Studi di Udine within the CRUI-CARE Agreement. This research was funded by the European Union, the European Regional Development Fund and the cross-border programme Interreg V-A Italy-Austria 2014–2020 (“RE-Cereal”, project number ITAT1005).
Author information
Authors and Affiliations
Contributions
GM designed the SPET experiment, performed data analysis and generated the scripts for plotting graphs and figures; SF managed breeding operations and data acquisition; SC, OP and VC designed the breeding scheme, DKS generated F1 hybrids, MM and GDG interpreted the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Serena Foria, Silvano Ciani, Ombretta Polenghi and Virna Cerne are employees of a food company seeking to develop proprietary broomcorn millet varieties.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Magris, G., Foria, S., Ciani, S. et al. Targeted sequencing of the Panicum miliaceum gene space and genotyping of variant sites from population genetics studies, combined in a single assay, as a tool for broomcorn millet assisted breeding. Euphytica 219, 102 (2023). https://doi.org/10.1007/s10681-023-03228-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03228-8