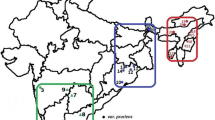
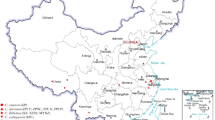
Abstract
Rhododendron canescens (Michaux) Sweet is a deciduous azalea from the southeastern United States that is used as an ornamental landscaping plant. We identified and characterized allelic variation in R. canescens architecture genes as the first step towards breeding a more compact phenotype for urban settings. The transcriptome of R. canescens vegetative and reproductive tissues was sequenced and analyzed using PacBio Iso-Seq methods. The analysis generated 24,244 full-length isoform sequences, of which 16,825 were annotated. Orthologs were identified of genes that have been shown to control height or branching across multiple plant species. These included genes for regulatory factors and components of phytohormone biosynthesis and signaling pathways. Exons of these genes were captured and sequenced from DNA libraries of 216 R. canescens plants, including a dwarf genotype. In this germplasm collection, 40 non-synonymous single-nucleotide polymorphisms (nsSNPs) were identified in R. canescens orthologs of GA INSENSITIVE (GAI), PHOTOPERIOD RESPONSIVE1 (PHOR1), GA-INSENSITIVE DWARF1 (GID1), BRASSINOSTEROID INSENSITIVE1 (BRI1), FLOWERING LOCUS C (FLC), MORE AXILLARY BRANCHING2 (MAX2), and BRANCHED1 (BRC1). The effect of amino acid changes on the primary and tertiary protein structure was examined in silico and several nsSNPs were found to be deleterious to protein function. Missense mutations were identified in RcMAX2 of the dwarf genotype. Discriminant Analysis of Principal Components found that RcMAX2 could be responsible for the difference between the dwarf and other R. canescens accessions. This investigation provides a pathway for breeding R. canescens with architectural traits better suited for urban environments.


Similar content being viewed by others
Data availability
All sequence data supporting this work are available in the NCBI SRA database under the project ID PRJNA685847.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Amador V, Monte E, García-Martínez JL, Prat S (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106:343–354
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33:2583–2585
Beveridge CA (2000) Long-distance signaling and a mutational analysis of branching in pea. Plant Growth Reg 32:193–203
Biemelt S, Tschiersch H, Sonnewald U (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Phys 135:254–265
Bolger AM, Marc L, Bjoern U (2014) A flexible trimmer for Illumina sequence data. Bioinformatics 15:2114–2120
Braun EM, Tsvetkova N, Rotter B, Siekmann D, Schwefel K, Krezdorn N, Plieske J, Winter P, Melz G, Voylokov AV, Hackauf B (2019) Gene expression profiling and fine mapping identifies a gibberellin 2-oxidase gene co-segregating with the dominant dwarfing gene ddw1 in rye (Secale cereale L.). Front Plant Sci 10:857
Bromberg Y, Rost B (2009) Correlating protein function and stability through the analysis of single amino acid substitutions. BMC Bioinform 10:1–9
Busov VB, Brunner AM, Strauss SH (2008) Genes for control of plant stature and form. New Phytol 177:589–607
Cantín CM, Arús P, Eduardo I (2018) Identification of a new allele of the Dw gene causing brachytic dwarfing in peach. BMC Res Notes 11:1–5
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33:W306–W310
Chamberlain D, Hyam R, Argent G, Fairweather G, Walter KS (1996) The genus Rhododendron: its classification and synonymy. Royal Botanic Garden, Edinburgh
Chappell M, Robacker C, Jenkins TM (2008) Genetic diversity of seven deciduous azalea species (Rhododendron spp. section Pentanthera) native to the eastern United States. J Am Soc Hortic Sci 133:374–382
Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22a-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10:231–243
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747
Clouse SD (2002) Brassinosteroid signal transduction: clarifying the pathway from ligand perception to gene expression. Mol Cell 10:973–982
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Dasgupta MG, Dharanishanthi V, Agarwal I, Krutovsky KV (2015) Development of genetic markers in Eucalyptus species by target enrichment and exome sequencing. PLoS ONE 10:e0116528
Fu Y, Springer NM, Gerhardt DJ et al (2010) Repeat subtraction-mediated sequence capture from a complex genome. Plant J 62:898–909
Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, Bouwmeester H (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194
Gonzalez-Garay ML (2016) Introduction to isoform sequencing using Pacific Biosciences technology (Iso-Seq). In: Wu J (ed) Transcriptomics and gene regulation. Springer, Dordrecht, pp 141–160
Götz S, García-Gómez JM, Terol J et al (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435
Günther T, Schmid KJ (2010) Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice. Theor Appl Genet 121:157–168
Guo J-C, Fang S-S, Wu Y et al (2019) CNIT: a fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res 47:W516–W522
Guo W, Chen L, Herrera-Estrella L, Cao D, Tran LSP (2020) Altering plant architecture to improve performance and resistance. Trends Plant Sci 25:1154–1170
Hedden P (2003) The genes of the Green Revolution. Trends Genet 19:5–9
Hill JL Jr, Hollender CA (2019) Branching out: new insights into the genetic regulation of shoot architecture in trees. Curr Opin Plant Biol 47:73–80
Jia X, Tang L, Mei X, Liu H, Luo H, Deng Y, Su J (2020) Single-molecule long-read sequencing of the full-length transcriptome of Rhododendron lapponicum L. Sci Rep 10:1–11
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Kim H, Tai TH (2019) Identifying a candidate mutation underlying a reduced cuticle wax mutant of rice using targeted exon capture and sequencing. Plant Breed Biotechnol 7:1–11
Kono TJ, Lei L, Shih CH, Hoffman PJ, Morrell PL, Fay JC (2018) Comparative genomics approaches accurately predict deleterious variants in plants. G3 Genes Genomes Genet 8:3321–3329
Kron KA, Gawen LM, Chase MW (1993) Evidence for introgression in azaleas (Rhododendron; Ericaceae): chloroplast DNA and morphological variation in a hybrid swarm on Stone Mountain. Georgia Am J Bot 80:1095–1099
Kurashige Y, Etoh JI, Handa T, Takayanagi K, Yukawa T (2001) Sectional relationships in the genus Rhododendron (Ericaceae): evidence from matK and trnK intron sequences. Plant Syst Evol 228:1–14
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:D458–D460
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin & Subgroup GPDP R (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079
Li A, Zhang J, Zhou Z (2014) PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform 15:311
Li XW, Zhu YL, Chen CY, Geng ZJ, Li XY, Ye TT, Mao XN, Du F (2020) Cloning and characterization of two chlorophyll A/B binding protein genes and analysis of their gene family in Camellia sinensis. Sci Rep 10:1–9
Liang YC, Reid MS, Jiang CZ (2014) Controlling plant architecture by manipulation of gibberellic acid signalling in petunia. Hort Res 1:1–6
Liu N, Zhang L, Zhou Y, Tu M, Wu Z, Gui D, Ma Y, Wang J, Zhang C (2021a) The rhododendron plant genome database (RPGD): a comprehensive online omics database for Rhododendron. BMC Genomics 22:1–10
Liu Q, Liaquat F, He Y, Munis MFH, Zhang C (2021b) Functional annotation of a full-length transcriptome and identification of genes associated with flower development in Rhododendron simsii (Ericaceae). Plants 10:649
Marroni F, Pinosio S, Di Centa E, Jurman I, Boerjan W, Felice N, Cattonaro F, Morgante M (2011) Large-scale detection of rare variants via pooled multiplexed next-generation sequencing: towards next-generation Ecotilling. Plant J 67:736–745
Moraes TS, Dornelas MC, Martinelli AP (2019) FT/TFL1: calibrating plant architecture. Front Plant Sci 10:97
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Muhr M, Prüfer N, Paulat M, Teichmann T (2016) Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED 1 expression and shoot architecture. New Phytol 212:613–626
Ng PC, Henikoff S (2006) Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet 7:61–80
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105–109
Savojardo C, Fariselli P, Martelli PL, Casadio R (2016) INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics 32:2542–2544
Scheiber SM, Jarret RL, Robacker CD, Newman M (2000) Genetic relationships within Rhododendron L. section Pentanthera G. Don based on sequences of the internal transcribed spacer (ITS) region. Sci Hort 85:123–135
Schiessl S, Samans B, Hüttel B, Reinhard R, Snowdon RJ (2014) Capturing sequence variation among flowering-time regulatory gene homologs in the allopolyploid crop species Brassica napus. Front Plant Sci 5:404
Silva-Junior OB, Grattapaglia D, Novaes E, Collevatti RG (2018) Design and evaluation of a sequence capture system for genome-wide SNP genotyping in highly heterozygous plant genomes: a case study with a keystone Neotropical hardwood tree genome. DNA Res 25:535–545
Sun L, Luo H, Bu D et al (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41:e166–e216
Sundaramoorthy J, Park GT, Jo H, Lee JD, Seo HS, Song JT (2021) A novel pinkish-white flower color variant is caused by a new allele of flower color gene W1 in wild soybean (Glycine soja). Agron 11:1001
Teichmann T, Muhr M (2015) Shaping plant architecture. Front Plant Sci 6:233
Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200
Vanholme B, Cesarino I, Goeminne G, Kim H, Marroni F, Van Acker R, Vanholme R, Morreel K, Ivens B, Pinosio S, Morgante M (2013) Breeding with rare defective alleles (BRDA): a natural Populus nigra HCT mutant with modified lignin as a case study. New Phytol 198:765–776
Vashisth T, Johnson LK, Malladi A (2011) An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep 30:2167–2176
Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59:253–279
Wang Y, Robacker CD, Braman SK (1998) Identification of resistance to azalea lace bug among deciduous azalea taxa. J Am Soc Hortic Sci 123:592–597
Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X (2013) Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 27:681–688
Wickham H, Averick M, Bryan J, Chang W, McGowan LDA, François R, Grolemund G, Hayes A, Henry L, Hester J (2019) Welcome to the Tidyverse. J Open Source Softw 4:1686
Yadav LK, McAssey EV, Wilde HD (2019) Genetic diversity and population structure of Rhododendron canescens, a native azalea for urban landscaping. HortSci 54:647–651
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591–1605
Zawaski C, Ma C, Strauss SH, French D, Meilan R, Busov VB (2012) PHOTOPERIOD RESPONSE 1 (PHOR1)-like genes regulate shoot/root growth, starch accumulation, and wood formation in Populus. J Exp Bot 63:5623–5634
Zhao J, Wang T, Wang M, Liu Y, Yuan S, Gao Y, Yin L, Sun W, Peng L, Zhang W, Wan J (2014) DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol 55:1096–1109
Zhao M, Chen P, Wang W, Yuan F, Zhu D, Wang Z, Ying X (2018) Molecular evolution and expression divergence of HMT gene family in plants. Int J Mol Sci 19:1248
Acknowledgements
The authors thank the Georgia Genomics and Bioinformatics Core (Athens, Georgia, USA) for technical support, Dr. Ron Miller for providing the dwarf R. canescens, and the USDA (Specialty Crop Block Grant 16-SCBGP-GA-0010; Hatch project GEO00755) for financial support.
Funding
This work was supported by USDA Specialty Crop Block Grant 16-SCBGP-GA-0010 and Hatch project GEO00755.
Author information
Authors and Affiliations
Contributions
LKY and HDW designed the experiment. LKY performed the experiment and analyzed the results. LKY and HDW wrote the manuscript and both authors read and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yadav, L.K., Wilde, H.D. Identification and bioinformatic characterization of rare variants of Rhododendron canescens architecture genes. Euphytica 218, 66 (2022). https://doi.org/10.1007/s10681-022-03019-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-03019-7