Abstract
Caraway (Carum carvi) is an economically important spice and medicinal plant of the Apiaceae family (syn. Umbelliferrae). Farmers often favor annual cultivation of caraway. However, the annual varieties, which are currently available, do not provide satisfying seed yields. Employing heterosis can be a promising approach to increase yield. Breeding of synthetic varieties utilizes heterosis and may be the method of choice for future caraway breeding. Knowledge of the outcrossing rate is important to evaluate the effectiveness of this breeding method. However, the outcrossing rate of caraway is unknown so far. We estimated the outcrossing rate of seven inbred lines under field conditions in a neighbor-balanced polycross design. For this purpose, we implemented a high-throughput genotyping system (PACE), accompanied by a high-throughput method for DNA extraction adapted to caraway. In total, more than 1300 individual plants were genotyped. We found a high variability of lines regarding outcrossing rate and other traits associated with flowering. The outcrossing rate was on average 66.5% and ranged from 51.6 to 82%. We discussed implications of our findings on the targeted breeding method.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Caraway (Carum carvi L., 2n = 2x = 20 (Pank 2012)) is a spice and medicinal plant of the Apiaceae family (syn. Umbelliferrae). Seeds are rich in essential oil, whose major components are limonene and carvone (Ruszkowska 1998). The European Pharmacopeia (Ph. Eur.) demands a minimum essential oil content of 30 ml/kg for pharmaceutical use (Ph. Eur. 2020). Originally, only biennial varieties were grown in Europe until annual varieties were introduced in the 1990s (Németh 1998). Potentially, annual varieties can substantially decrease production costs and thus are of high interest for breeders and farmers (Pank 2012). In the beginning, annual varieties had a low essential oil content (Pank 2012). ‘Sprinter’ was the first annual variety which fulfilled the requirements of Ph. Eur. (Pank 2012). However, annual varieties do not gain satisfying seed yields compared to biennial varieties so far (Erpenbach 2012).
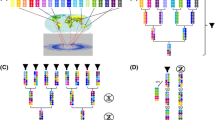
In many crops, breeding achieved major yield increases by exploiting heterosis (Liu et al. 2020). Hybrid breeding is the breeding method that exploits heterosis most intentionally. However, hybrid breeding requires an efficient system to control pollination (e.g. cytoplasmic male sterility). If such a system is not available, breeding of synthetic varieties can be an alternative method to exploit heterosis. This method was summarized by Becker (1988): It was classified as a specialized method of population breeding. The major difference to hybrid breeding is uncontrolled open pollination. According to Becker (1988), synthetic varieties differ from usual population varieties in three aspects: “parents of a synthetic variety are selected due to general combining ability (GCA)”, “parents are maintained” and “the number of parents [..] is usually very restricted”. Briefly, breeding material (e.g. inbred lines) is placed in a testcross (polycross or topcross) to produce seeds by random pollination. The testcross is followed by a yield test of the outcrossed material to estimate GCA. The best lines are selected and crossed again (Syn0). Seeds from this crossing (Syn1) or further propagated seeds (Syn2−x) constitute a synthetic variety. Figure 1 outlines the breeding scheme.
A conceivable concept of a breeding scheme to produce synthetic varieties in caraway (modified after Becker (2011)). Inbred lines and their progenies are indicated by different colors
As an efficient system to control pollination is lacking in caraway, breeding of synthetic varieties might be the method of choice to exploit heterosis in caraway. To the best of our knowledge, heterosis has not been systematically exploited in caraway. The only exception is the breeding of the winter-annual variety 'Aprim', which contained elements towards utilizing heterosis (Smirous, P. 2020, Agritec Ltd., personal communication). Pank et al. (2007) developed the inbred lines that were used in this study and Pank performed a polycross in a lower inbred generation (I3). However, no heterosis was detected in this early trial. Therefore, we do not know to what extent heterosis can be exploited in caraway. In conclusion from the mating system of caraway, investigations on heterosis should be worthwhile.
In general, heterosis is strong in outcrossing species (Liu et al. 2020) and caraway is assumed to predominately exhibit outcrossing. Caraway is visited by a variety of insect species (Langenberger and Davis 2002a, b), which promote pollination, but pollination by wind can play an additional role (Bouwmeester and Smid 1995). Caraway flowers are protandrous, i.e., male anthers mature and die back before female stigmata mature (Mihalik 1998; Pank 2012). Németh et al. (1999) showed in isolation experiments that self-fertilization within the same umbel is very low. Self-fertilization between umbels of the same plant within different stages is more likely, but still low. Considerably higher fertilization rates were observed after open pollination. However, measuring fertilization rates does not enable differentiation and quantification of self-fertilization and outcrossing in open pollination. For this purpose, F1 plants have to be analyzed using morphological or genetic markers.
Whether morphological markers (like flower or seed color) can be used, depends on the plant species and the varieties chosen for investigation. If no morphological traits are available, a multitude of genetic marker systems will be at disposal nowadays. Amplified fragment length polymorphism (AFLP) markers or simple sequence repeats (SSR) markers were often used for the estimation of the outcrossing rate (OCR) (Penzkofer et al. 2018; Santos and Gama 2013; Santos and Neto 2011; Sharma et al. 2017; Sinha et al. 2015). In contrast to AFLP markers, SSR markers require prior knowledge on DNA sequences (Garrido-Cardenas et al. 2018; Vieira et al. 2016). The development of next generation sequencing (NGS) methods led to an increasing use of single nucleotide polymorphisms (SNPs) (Garrido-Cardenas et al. 2018; Semagn et al. 2014). Among others, single SNPs can be re-identified using polymerase chain reaction (PCR) based marker systems like competitive allele-specific PCR (KASP, LGC) or PCR allele competitive extension (PACE, 3cr Bioscience). These systems consist of a PCR with an endpoint fluorescence measurement. Simplified, allele specific primers compete for annealing to the SNP sequence and release different fluorescent dyes if successful. KASP markers were widely used in recent plant scientific studies (Jatayev et al. 2017). However, we found no explicit application for OCR estimation in literature. Except, Cottage et al. (2012) used KASP markers to detect outcrossed individuals in an inbreeding scheme.
To date, only randomly amplified polymorphic DNA (RAPD) markers were used for genetic analysis of caraway (Bocianowski and Seidler-Lozykowska 2012; Laribi et al. 2011; Seidler-Lozykowska et al. 2014). A reference genome is not available for caraway. Recently, we conducted a genotyping by sequencing (GBS). This NGS method, developed by Elshire et al. (2011), led to the discovery of thousands of SNP loci, which potentially can be converted to a marker system like PACE or KASP.
Our general goal is to evaluate breeding of synthetic varieties as a method for caraway breeding. As a first step, we performed a testcross, implementing a neighbor-balanced polycross design. In this study, we evaluated the functionality of this design for caraway. We focused on the analysis of OCR because outcrossing is essential to exploit heterosis. To estimate OCR, we adopted the PACE genotyping system and an efficient DNA extraction protocol for a 96 samples scale.
Material and methods
Material
In the 1990s, biennial and annual material was crossed and progenies were selected towards annual flowering and high essential oil content. Beginning in the F2 generation, plants were coerced to self-fertilization using isolation bags. Finally, this resulted in inbred lines of the fourth (I4) or fifth (I5) inbred generation. Nine I4 lines and nine I5 lines from diverse pedigrees were selected to test breeding of synthetic varieties for caraway.
Field trials
A neighbor-balanced polycross design was fitted using the algorithm provided by Varghese et al. (2015), which implements the design according to Olesen (1976). This method provides polycross designs for “v genotypes balanced for neighbors (including self) in eight directions […] (with v + 1 as prime)” (Varghese et al. 2015). The design can be described as a specialized Latin square. In theory, nine Latin squares are necessary for a complete balanced design of 18 lines. However, merely three Latin squares could be realized, because seed availability was restricted. In May 2018, pre-grown plants in early leaf stage (BBCH 13–16 (Hack et al. 1992)) were planted with a distance of 50 cm between the plants. Each Latin square contained 18 plants per line, so that each line was represented by 54 plants in total (A picture is provided in Supplementary Figure S1). At maturity (brown seeds, BBCH = 89 (Hack et al. 1992)), single plants were harvested and pooled for each line. This resulted in 18 F1 populations.
We measured seed yield, thousand-grain weight (TGW) and germination rate for each pooled harvest. Three times 100 single seeds were counted and weighed (TGW). These seeds were sown in soil under greenhouse conditions. Germinated plants were counted 21 days after sowing. Based on these data, we estimated the number of potential field plots for GCA testing with a size of 12 m2 per plot and 100 germinating seeds per m2.
In parallel to the polycross, the lines were grown in separate small plots of 25 plants, to estimate flowering traits and plant height without disturbing the polycross. Up to three repetitions (L04 is missing due to reduced seed availability, Supplementary Table S1) per line were grown in a randomized complete block design. The begin of flowering was defined as the opening of the first umbel (BBCH = 60 (Hack et al. 1992)) for each single plant. For this study, we computed the 0.1, 0.5 and 0.9 quantile for each plot (= 10%, 50% and 90% of plants in a plot are flowering). The end of flowering was defined as the decay of petals in the last flowering umbels (BBCH = 67 (Hack et al. 1992)). The end of flowering of a plot was defined as the time point, at which more than 90% of plants per plot finished flowering. Plant height was measured at the end of flowering.
Both field trials were planted at the JKI location in Quedlinburg, Saxony-Anhalt. Irrigation was carried out after planting. The year 2018 was an exceptional hot and dry year. In particular, places in Saxony-Anhalt suffered from severe drought from February until November (Deutscher Wetterdienst, “The weather in Germany in 2018”, Pressemitteilung, Offenbach, 2018-12-28). In total, a precipitation of 383 mm/m2 was measured at the JKI location in Quedlinburg. Supplementary Figure S2 provides an overview of the weather at the JKI location in 2018.
DNA extraction
A protocol for DNA extraction in barley (Brandes, in prep.) was slightly modified for DNA extraction in caraway. 192 plants per each F1 population (n = 7) were grown in greenhouse in 96 pot plates. The youngest developing leaf per plant (BBCH = 15–18 (Hack et al. 1992)) was sampled. Leaves were collected in racked collection microtubes (Cat. no. 19560, Qiagen). Two 4 mm steel beads per sample were added. Samples were frozen with liquid nitrogen. Samples were ground using mixer mill MM 300 (Retsch). Samples were incubated at 65 °C for 45 min after adding 600 µl extraction buffer (1 M guanidine thiocyanate, 2 M NaCl, 30 mM NaAc) per sample. Samples were spun using Rotina 420 centrifuge (Hettich) for 30 min at 4000 rpm. 400 µl of liquid per sample were transferred to new collection microtubes and 2.5 µl RNase A (5 mg/ml) was added. Samples were incubated for 30 min at 35 °C. DNA was precipitated, adding 300 µl propan-2-ol per sample. Samples were spun using Rotina 420 centrifuge (Hettich) for 30 min at 4000 rpm. DNA pellets were washed in two steps, adding 600 µl per solution and sample (wash solution 1: 76% ethanol, 200 mM NaAc; wash solution 2: 76% ethanol, 10 mM NaAc). Dried DNA was dissolved in 600 µl TE-buffer (10 mM Tris/HCl, 1 mM EDTA) per sample. DNA was quantified using nanodrop 8000 spectrophometer (Thermo Fisher Scientific).
Marker design and marker analysis
In 2018, GBS analysis and SNP calling were conducted including all 18 inbred lines used in this study. Because a reference genome was not available, a clustering was carried out, to which we refer as a reference. To estimate OCR for a specific line, the resulting dataset from SNP calling was searched for SNPs that showed one homozygous genotype for the specific line (the maternal line) and the other homozygous genotype for the other 17 lines (potential paternal lines). Then, the percentage of heterozygous genotypes in a F1 population corresponds with OCR. This search was carried out for each line as a maternal line. Selected SNPs were transferred to the co-dominant PACE genotyping system (3cr Bioscience). Flanking SNP sequences, for which marker design was successful, are listed in Table 1.
In detail, the PACE genotyping master mix (standard ROX) was used (3cr Bioscience). The service of 3cr Bioscience included the computation of primer sequences. DNA primers were produced by Invitrogen (Thermo Fisher Scientific). For each marker, common primer (30 µM) and both allele specific primers (12 µM) were composed to an assay mix. Genotyping was carried out according to the manufacturer's protocol, except that reaction volume was reduced from 10 µl to 5 µl. One PCR reaction contained 2.43 µl of master mix, 0.07 µl of assay mix and 2.5 µl of sample DNA of variable concentration. PCR, endpoint fluorescence measurement and data processing were carried out using the C1000 Touch thermal cycler, including the CFX96 real-time system and the CFX Maestro software (Bio-Rad). Thermal cycling conditions are listed in Table 2. If necessary to determine the genotype reliably, ten additional cycles were added (20 s 94 °C and 60 s at 57 °C).
Designed markers were tested for functionality with DNA samples of all 18 lines (pools of five individual plants per line), two designed heterozygous genotypes (mixture of maternal line and a potential paternal line in equal amounts) and two negative controls. In total, 20 markers were tested, but only seven markers could be used for OCR estimation (Table 3). These seven markers enabled the estimation of OCR within seven F1 populations, which originated from seven different maternal lines. Each PCR run contained two samples of the maternal line, two samples of potential paternal lines (positive controls) and two negative controls.
Statistics
Statistical analyses and visualization of the phenotypic data were carried out by using R software (R Core Team 2018) and the R package ggplot2 (Wickam 2016). Pearson correlation coefficients were determined by using the function rcorr implemented in the R package Hmisc (v.4.3-1, Harrell 2020).
Results
Marker functionality tests, DNA extraction and marker analysis
20 PACE markers were designed to estimate OCR in the set of 18 inbred lines under investigation. Seven markers showed the desired separation of genotypes. These markers could be used for OCR estimation. In detail, a functional marker showed the homozygous alternative genotype for the maternal line (in this case always a predominant HEX signal) and the homozygous reference genotype for the 17 potential paternal lines (in this case always a predominant FAM signal). For the two mixed DNA samples (maternal and paternal DNA in equal amounts), a functional marker showed a heterozygous genotype (mixed HEX and FAM signal).
Five markers showed allelic discrimination, but not the desired genotype for all 18 lines. Four else markers showed amplification only for one of the two alleles. Four markers were non-functional, i.e., no considerable amplification took place. Therefore, in 35% of the cases, SNP genotypes could be confirmed for all 18 lines using PACE. In 60% of the cases, at least the presence of a SNP was confirmed.
In a test (n = 96), the DNA concentration ranged from 31 to 254 ng/µl (mean 103.26 ng/µl ± 44.62 ng/µl SD). No standardization of the concentration was necessary before genotyping. To determine the genotype reliably, a few samples needed ten additional PCR cycles. In total, 98.66% of the samples could be genotyped successfully. At least 185 samples per examined maternal line were successfully genotyped.
Outcrossing rate (OCR)
Figure 2 exemplary demonstrates the output of a PACE assay for 96 samples. We found a considerable variability for OCR between the investigated seven F1 populations (Fig. 3). The proportion of heterozygous genotypes (= OCR) ranged from 51.6 to 82% and averaged 66.5% (± 11% SD). Vice versa, the proportion of homozygous maternal genotypes ranged from 15.3 to 47.9% (on average 32.4% ± 11.3% SD). On average, 1.1% (± 1.3% SD) of F1 plants were found to be homozygous paternal genotypes (at maximum 3.2%).
Other traits and correlations
A summary of all investigated traits is shown in Table 4. Detailed information is provided in Supplementary Table S1. In general, variability was high between lines for several traits. In detail, the begin of flowering (quantile 0.5) ranged from 71 to 104 days after sowing. The seed yield per line ranged from 264.9 to 1495.6 g. In detail, we observed the flowering period of the lines. Figure 4 displays the period from begin of flowering (quantile 0.1) to end of flowering (quantile 0.9). There was an overlapping flowering period between all lines, but altogether the flowering period considerably differed between some lines. The germination rate of F1 populations was generally low.
Pearson correlation coefficient was determined to reveal correlations between traits, for n = 7, if OCR is included and n = 17, if OCR is excluded. The begin of flowering is highly correlated (p < 0.001) with the end of flowering, i.e., most early flowering lines also finished flowering early (Table 5). Furthermore, the begin and the end of flowering were significantly negatively correlated with seed yield (p < 0.05/0.01) and germination rate (p < 0.01), i.e., early flowering promoted seed yield and germination ability. In addition, the begin of flowering (BoF Q 0.9) tended to be negatively correlated with OCR, but this correlation was not significant (p = 0.29, Table 5).
Discussion
General remarks
As a first step towards breeding synthetic varieties in caraway, we conducted a polycross. The sole purpose of the polycross is to produce F1 seeds, which are sown in yield tests to estimate GCA for each inbred line. However, seed production within a polycross might not be as trivial as assumed at first glance. In theory, the polycross should promote completely random pollination (Varghese et al. 2015). Obviously, simultaneous flowering of all plants is necessary to reach this goal (Varghese et al. 2015). Ideally, each plant disperses the same amount of pollen in each direction at the same time. Then, we would achieve panmixis in a polycross. Definitely, we will never reach this ideal situation in a practical breeding approach. Several studies on various species found unequal distribution of paternity after mating in a polycross (Moriguchi et al. 2009; Norman et al. 2020; Riday et al. 2013; Tew and Pan 2010). We evaluated how far we missed completely random pollination in the conducted polycross. Subsequently, we will discuss the implications of our findings on the breeding of synthetic varieties as a method for caraway breeding. In addition, we will discuss the functionality of the implemented genotyping system. Prior to all of that, we will evaluate the estimation of OCR and other traits per se with the purpose to highlight validity and limitations of our findings.
OCR, other traits and correlations—validity and limitations
In general, the used marker set did not allow specific determination of paternity. We could only distinguish between outcrossing (= fertilization by one of the 17 other lines) and self-fertilization. More precisely, we included fertilization between different individuals of the same line into the term "self-fertilization". To a small extent, we found homozygous paternal genotypes in F1 populations. In the event that neighboring plants grew or fell into each other and were incompletely separated during the harvest, contaminations might occur. The distance of 50 cm between plants usually prevented extensive intermingling between neighboring plants. We assume that the maximum impurity of 3.2%, which was detected in this study, is acceptable and should not largely influence GCA testing. Minor allele frequency of the paternal allele in a maternal line could be another explanation for homozygous paternal genotypes. Since homozygosity is not complete in I4 or I5 inbred lines, we cannot exclude this. If this is the case, a small number of heterozygous genotypes can actually result from self-fertilization. To conclude, we cannot be certain on the origin of the homozygous paternal genotypes. For simplicity, we equated the percentage of heterozygous genotypes with OCR and did not exclude homozygous paternal genotypes from analysis.
With a minimum OCR of 51.6% and an average OCR of 66.5%, we can state that outcrossing dominates in caraway, but self-fertilization can play a considerable role. Mixed mating systems were frequently observed in various plant species (Whitehead et al. 2018). Outcrossing rates were measured for some other species of the Apiaceae family: Gaudeul and Till-Bottraud (2003) found an OCR from 65 to 100% in Eryngium alpinum in natural populations. Rong et al. (2010) found an average OCR of 96% for wild Daucus carota. Several species of the Apiaceae are protandrous (Schlessman et al. 2004; Webb 1981), which might promote predominant but incomplete outcrossing. Therefore, our findings on caraway could be typical for several Apiaceae species.
In general, our findings on OCR are limited to the examined genotypes and to the certain year and location of growing. All tested genotypes are inbred lines. The inbreeding process might have selected genotypes with a high self-fertilization ability. Therefore, OCR might be higher in (natural) populations. In this context, we may mention that we never observed complete self-incompatibility during propagations using isolation bags, if wild accessions or cultivars from population breeding were propagated. The year 2018 had an unusual dry and hot spring and summer, which might have reduced pollen viability. “Male gametophyte development has been identified as the most heat-vulnerable stage” in many plants (Lohani et al. 2020). Reduced pollen viability decreases fertilization rates, but it is less certain in which direction drought and heat stress would affect OCR. Bishop et al. (2017) found that OCR increased under heat stress in Vicia faba. To some extent, this insect pollinated species might be comparable to caraway. We know that a variety of insects visit caraway (Langenberger and Davis 2002b), but we do not know which species are most important for pollination. Therefore, we cannot draw conclusions for optimal pollination conditions. Anyway, higher pollinator abundance must not necessarily enhance outcrossing. Due to protandry (Mihalik 1998; Pank 2012), self-fertilization requires pollen transfer as well, so that higher pollinator abundance should likewise promote self-fertilization. The possibility of partial wind pollination (Bouwmeester and Smid 1995) adds further uncertainty.
Altogether, OCR has to be estimated in multiple years and on multiple locations to determine environmental effects on OCR in caraway. However, a polycross is a laborious task, which cannot be conducted in each breeding year. In addition, most of the tested 18 inbred lines will not play any role in further breeding. From a breeder's perspective, it is of more importance to estimate OCR again between the few lines that will be selected for breeding a synthetic variety. We endeavor to accompany further breeding processes by genotypic evaluation.
Evaluation of the polycross design regarding random pollination
First, random pollination is limited by the number of manageable replications of Latin squares of the polycross design. Based on the algorithm of Varghese et al. (2015), nine different Latin squares would be necessary to complete the neighbor-balanced polycross design. However, nine Latin squares are scarcely manageable and seeds were not available in a sufficient amount. Nevertheless, we planted an unusual high number of plants with three Latin squares and in total 54 plants per line. Becker (2011) states that usually ten to 30 plants per line are planted in a testcross. We produced a sufficient amount of seeds for at least two years of GCA testing (with four repetitions each season) despite a low germination rate of F1 seeds. (We assume this low germination rate was caused by severe drought and is no inherent problem of the polycross.) Finally, we assume that we made a good compromise between enabling random pollination and practicability in a breeding program.
High variability between the inbred lines regarding flowering period and OCR might have reduced random pollination stronger than the reduced number of polycross replications. All lines had an overlapping flowering period to some extent, but lines largely differed in begin of flowering and end of flowering. Therefore, lines had different peaks of flowering and periods occurred in which only a subset of lines could pollinate each other. In addition, it is necessary to take into account that differences in pollination ability between lines can exist. Future studies may also investigate pollen amounts and pollen viability. To summarize, completely random pollination was not given in the polycross design in this study due to a variability of inbred lines regarding flowering traits. Nevertheless, this will not strongly influence the production of F1 seeds for GCA testing, but we should discuss the implications of our findings with regard to the breeding strategy.
Implications of findings on the breeding method
First, we confirmed that caraway is predominately an outcrossing species. From an outcrossing species, we might expect heterosis in F1 populations produced in a polycross. However, the so-called F1 populations can still contain self-fertilized plants of the corresponding inbred line to a considerable amount. Therefore, potential heterosis will not entirely be exploited in GCA testing.
The found large range in OCR between lines (51.6 to 82%) suggests that OCR could have a considerable effect on the combining ability of the lines. We assume that a higher OCR positively affects heterosis and hence the combining ability of a line. Vice versa, selecting for high GCA could lead to a selection for high OCR. This seems to be an advantageous situation for the breeding process. However, we have to consider that the OCR of a line is probably not a constant. The OCR of a line might largely depend on the certain set of lines, in which a line is tested. As shown, lines can vary in flowering time. In addition, lines might vary in pollination ability based on differences in the quantity of produced viable pollen. The latter remains to be investigated. In this light, selection decisions have to be carefully conducted.
-
1.
Flowering time Differences in begin and end of flowering could cause severe problems, if not properly considered in selection decisions. Obviously, we only observe heterosis between lines that are actually crossed in the polycross. Therefore, if lines with high GCA but different flowering periods are combined, we might not achieve the expected heterosis because outcrossing would not take place between all components. Ideally, reliable data for begin and end of flowering of lines are already available from pretests before testcrossing. Then, subsets of lines with similar flowering periods can be composed and placed in separate testcrosses. If, as in our case, time for thorough pretests is not available, combining of lines with different flowering traits should at least be avoided for the composition of a synthetic variety.
-
2.
Pollination ability It is possible that some lines merely produce a low quantity of viable pollen and are less able to fertilize other lines and likewise to self-fertilize. Therefore, such lines might show a higher OCR. Low pollen production might be associated with a high degree of inbreeding depression. Under these assumptions, lines with high inbreeding depression might be able to show a high positive GCA due to high OCR. Then, selecting merely based on GCA might lead to erroneous selection decisions. Firstly, lines whose high GCA is mainly attributable to high OCR would depend on good pollen donators, which might not be selected for Syn1 production. Secondly, the potential heterosis of lines with a high yield performance per se might be underestimated. Such lines might have a higher OCR and might show higher heterosis, if placed in a set of lines that all produce high amounts of viable pollen. In our study, the lines with the lowest (L18) and highest (L02) seed yield in the polycross (and with an available OCR estimate) both exhibited a low OCR (51.6% and 56.2%, respectively). That speaks against the theory raised above, but statistical evidence is yet low. We conclude that it will be important to estimate not only the yield of the F1 populations, but also the yield of the inbred lines per se and to analyze the correlations between OCR, GCA and the yield of the inbred lines per se.
The incomplete outcrossing in the polycross suggests that maximum possible heterosis will not be achieved in Syn1 generation. It should be tested, whether heterosis can be increased in further generations (Syn2 or Syn3), if outcrossing proceeds. However, OCR might be higher during Syn1 production than the estimated average of 66.5% in the polycross: Firstly, selection based on GCA might favor lines with high OCR. Secondly, Syn1 production does not take place in a polycross, but by sowing a mixture of selected lines. Plants will grow much closer, which could improve outcrossing. Vantreuren et al. (1993), for instance, showed a positive correlation between plant density and OCR in Salvia pratensis. Thirdly, lines should be better selected for simultaneous flowering. After GCA testing, genotypic and phenotypic investigations on Syn1 production should be the next step in evaluating the breeding method.
Evaluation of the PACE genotyping system and DNA extraction protocol
Finally, we may also discuss the PACE genotyping system used for OCR estimation. As a codominant marker system, PACE allowed the distinction of the two homozygous genotypes (maternal and paternal) and the heterozygous (outcrossed) genotype in a biallelic context. As one advantage (compared to the rather dominant AFLP marker system (Semagn et al. 2006)), we could detect some probable contaminations. The success rate of 35% was rather low for conversion of a SNP from GBS to the PACE genotyping system. Generously, we could state a success rate of 60%, if we take all markers into account that confirmed the presence of a SNP. Partially, the rather low success rate in our study might result from the special trial design. The allele of the maternal line was often a very seldom allele so that the probability was high to address an erroneous SNP.
The adapted protocol for DNA extraction was highly efficient. To the best of our knowledge, it is the first published protocol for caraway which is suitable for extraction of 96 samples in a single run. Depending on downstream application, some reductions of protocol steps (RNase treatment and washing) can be tested to reduce working time. We assume that PACE accompanied by this protocol can be a very efficient genotyping system for future genetic analysis of caraway.
Conclusions
We assume that the applied neighbor-balanced polycross design should work to produce F1 seeds for GCA testing in caraway. However, we found that high variability in flowering traits reduced the ideally endeavored random pollination. With an average OCR of 66.5%, we can confirm that caraway is predominately an outcrossing species, but self-fertilization can take place to a considerable amount. The mating system might be best described as a mixed mating system. Additional years, locations and genotypes should be investigated to get deeper insights into environmental and genotypic effects on OCR. Based on our findings on OCR, we expect to find heterosis in F1 generations. As a next step, GCA testing should be conducted to show to what extent heterosis can be exploited in caraway. Since we detected considerable differences in OCR between lines, OCR could have a major effect on the combining ability of a line. The correlations between OCR, GCA and the yield of the inbred lines per se should be thoroughly investigated. For the first time, we implemented PACE as a high-throughput genotyping system accompanied by an efficient DNA extraction protocol for caraway. Both should be of use in several future genetic studies.
Data availability
Summary data, relevant sequence and SNP data used for this study are included in the manuscript or in supplementary material.
Code availability
No specialized code was used in this study.
References
Becker HC (1988) Breeding synthetic varieties of crop plants. Plant Genet Breed Rev 31–54
Becker HC (2011) Pflanzenzüchtung. 2 edn. Ulmer UTB
Bishop J, Jones HE, O’Sullivan DM, Potts SG (2017) Elevated temperature drives a shift from selfing to outcrossing in the insect-pollinated legume, faba bean (Vicia faba). J Exp Bot 68:2055–2063. https://doi.org/10.1093/jxb/erw430
Bocianowski J, Seidler-Lozykowska K (2012) The relationship between RAPD markers and quantitative traits of caraway (Carum carvi L.). Ind Crops Prod 36:135–139. https://doi.org/10.1016/j.indcrop.2011.08.019
Bouwmeester HJ, Smid HG (1995) SEED YIELD IN CARAWAY (CARUM-CARVI).1. ROLE OF POLLINATION. J Agric Sci 124:235–244. https://doi.org/10.1017/s0021859600072907
Cottage A, Gostkiewicz K, Thomas JE, Borrows R, Torres AM, O’Sullivan DM (2012) Heterozygosity and diversity analysis using mapped single nucelotide polymorphisms in a faba bean inbreeding programme. Mol Breed 30:1799–1809. https://doi.org/10.1007/s11032-012-9745-4
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (gbs) approach for high diversity species. PLoS ONE 6:10. https://doi.org/10.1371/journal.pone.0019379
Erpenbach JT (2012) Pflanzenbauliche und züchterische Beurteilung von ein- und zweijährigen Kümmelsorten und -herkünften (Carum carvi L.). University of Bonn
Garrido-Cardenas JA, Mesa-Valle C, Manzano-Agugliaro F (2018) Trends in plant research using molecular markers. Planta 247:543–557. https://doi.org/10.1007/s00425-017-2829-y
Gaudeul M, Till-Bottraud I (2003) Low selfing in a mass-flowering, endangered perennial, Eryngium alpinum L. (Apiaceae). Am J Bot 90:716–723. https://doi.org/10.3732/ajb.90.5.716
Hack H, Bleiholder H, Buhr L, Meier U, Schnock-Fricke U, Weber E, Witzenberger A (1992) Einheitliche Codierung der phänologischen Entwicklungsstadien mono- und dikotyler Pflanzen – erweiterte BBCH-Skala, Allgemein. Nachrichtenblatt des deutschen Pflanzenschutzdienstes 44:265–270
Harrell FE (2020) Hmisc: Harrell Miscellaneous. R package version 4.3-1
Jatayev S et al (2017) Advantages of Amplifluor-like SNP markers over KASP in plant genotyping. BMC Plant Biol 17:11. https://doi.org/10.1186/s12870-017-1197-x
Langenberger MW, Davis AR (2002a) Honey bee pollen foraging in relation to flowering phenology of biennial caraway (Carum carvi L.). Can J Plant Sci 82:203–215. https://doi.org/10.4141/p01-019
Langenberger MW, Davis AR (2002b) Temporal changes in floral nectar production, reabsorption, and composition associated with dichogamy in annual caraway (Carum carvi; Apiaceae). Am J Bot 89:1588–1598. https://doi.org/10.3732/ajb.89.10.1588
Laribi B, Zoghlami N, Lamine M, Kouki K, Ghorbel A, Mougou A (2011) RAPD-based assessment of genetic diversity among annual caraway (Carum carvi) populations. Eurasian J Biosci 5:37–47
Liu J, Li MJ, Zhang Q, Wei X, Huang XH (2020) Exploring the molecular basis of heterosis for plant breeding. J Integr Plant Biol 62:287–298. https://doi.org/10.1111/jipb.12804
Lohani N, Singh MB, Bhalla PL (2020) High temperature susceptibility of sexual reproduction in crop plants. J Exp Bot 71:555–568. https://doi.org/10.1093/jxb/erz426
Mihalik E (1998) Taxonomy and botanical description of the genus Carum. In: Németh É (ed) Caraway The Genus Carum, vol 7. Medicinal and Aromatic Plants - Industrial Profiles. Harwood Academic Publishers, Amsterdam, pp 9–34
Moriguchi Y, Ishiduka D, Kaneko T, Itoo S, Taira H, Tsumura Y (2009) The contribution of pollen germination rates to uneven paternity among polycrosses of cryptomeria japonica. Silvae Genet 58:139–144. https://doi.org/10.1515/sg-2009-0018
Németh É (1998) Introduction. In: Nemeth E (ed) Caraway The Genus Carum, vol 7. Medicinal and Aromatic Plants - Industrial Profiles. Harwood Academic Publishers, Amsterdam, pp 1–8
Németh É, Bernáth J, Petheô F (1999) Study on flowering dynamic and fertilization properties of caraway and fennel. Acta Hortic 502:77–84. https://doi.org/10.17660/ActaHortic.1999.502.9
Norman PE et al (2020) Paternity assignment in white guinea yam (dioscorea rotundata) half-sib progenies from polycross mating design using SNP markers. Plants-Basel 9:15. https://doi.org/10.3390/plants9040527
Olesen K (1976) A completely balanced polycross design. Euphytica 25:485–488. https://doi.org/10.1007/BF00041582
Pank F (2012) Kümmel (Carum carvi L.). In: Hoppe B (ed) Handbuch des Arznei- und Gewürzpflanzenanbaus, vol 4. Verein für Arznei- und Gewürzpflanzen SALUPLANTA e.V., Bernburg, pp 728–762
Pank F, Kruger H, Quilitzsch R (2007) Results of a polycross-test with annual caraway (Carum carvi L. var. annuum hort). Zeitschrift Für Arznei- & Gewürzpflanzen 12:127–135
Penzkofer M, Seefelder S, Heuberger H (2018) Estimation of outcrossing rates using genomic marker and determination of seed quality parameters in Valeriana officinalis L. s.l. under field conditions. Euphytica 214:15. https://doi.org/10.1007/s10681-018-2164-9
Ph . Eur. (2020) Kümmel Carvi Fructus. Europäisches Arzneibuch 10:2270
R Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing
Riday H, Johnson DW, Heyduk K, Raasch JA, Darling ME, Sandman JM (2013) Paternity testing in an autotetraploid alfalfa breeding polycross. Euphytica 194:335–349. https://doi.org/10.1007/s10681-013-0938-7
Rong J, Janson S, Umehara M, Ono M, Vrieling K (2010) Historical and contemporary gene dispersal in wild carrot (Daucus carota ssp. carota) populations. Ann Bot 106:285–296. https://doi.org/10.1093/aob/mcq108
Ruszkowska J (1998) Main chemical constituents of Carum. In: Nemeth E (ed) Caraway the genus carum. Medicinal and Aromatic Plants - Industrial Profiles, Amsterdam, pp 35–54
Santos CAF, Gama R (2013) An AFLP estimation of the outcrossing rate of Spondias tuberosa (Anacardiaceae), an endemic species to the Brazilian semiarid region. Rev Biol Trop 61:577–582
Santos CAF, Neto FPL (2011) Outcrossing rate between “Haden” and “Tommy Atkins” mangoes estimated using microsatellite and AFLP markers. Pesquisa Agropecuaria Brasileira 46:899–904. https://doi.org/10.1590/s0100-204x2011000800016
Schlessman MA, Barrie FR, van Wyk BE, Tilney PM (2004) Protogyny in Apiaceae, subfamily Apioideae: systematic and geographic distributions, associated traits, and evolutionary hypotheses. S Afr J Bot 70:475–487. https://doi.org/10.1016/S0254-6299(15)30232-5
Seidler-Lozykowska K, Kuczynska A, Mikolajczyk K, Nowakowska J, Bocianowski J (2014) Estimation of genetic distance among genotypes of caraway (Carum carvi L.) using RAPD-PCR. Acta Sci Agron 36:183–188. https://doi.org/10.4025/actasciagron.v36i2.18197
Semagn K, Bjornstad A, Ndjiondjop MN (2006) An overview of molecular marker methods for plants. Afr J Biotechnol 5:2540–2568
Semagn K, Babu R, Hearne S, Olsen M (2014) Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed 33:1–14. https://doi.org/10.1007/s11032-013-9917-x
Sharma SS, Islam MA, Negi MS, Tripathi SB (2017) Estimation of outcrossing rates in biodiesel species pongamia pinnata based on AFLP and microsatellite markers. Natl Acad Sci Lett India 40:105–108. https://doi.org/10.1007/s40009-016-0533-2
Sinha P, Islam MA, Negi MS, Tripathi SB (2015) Estimation of outcrossing rates in interspecific backcross plants of Jatropha curcas (L.) using AFLP and SSR markers. Physiol Mol Biol Plants 21:605–609. https://doi.org/10.1007/s12298-015-0318-y
Tew TL, Pan YB (2010) Microsatellite (simple sequence repeat) marker-based paternity analysis of a seven-parent sugarcane polycross. Crop Sci 50:1401–1408. https://doi.org/10.2135/cropsci2009.10.0579
Vantreuren R, Bijlsma R, Ouborg NJ, Vandelden W (1993) The effects of population-size and plant-density on outcrossing rates in locally endangered salvia-pratensis. Evolution 47:1094–1104. https://doi.org/10.1111/j.1558-5646.1993.tb02138.x
Varghese C, Varghese E, Jaggi S, Bhowmik A (2015) Experimental designs for open pollination in polycross trials. J Appl Stat 42:2478–2484. https://doi.org/10.1080/02664763.2015.1043860
Vieira MLC, Santini L, Diniz AL, Munhoz CdF (2016) Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol 39:312–328. https://doi.org/10.1590/1678-4685-GMB-2016-0027
Webb CJ (1981) Andromonoecism, protandry, and sexual selection in umbelliferae. N Z J Bot 19:335–338. https://doi.org/10.1080/0028825x.1981.10426389
Whitehead MR, Lanfear R, Mitchell RJ, Karron JD (2018) Plant mating systems often vary widely among populations. Front Ecol Evol. https://doi.org/10.3389/fevo.2018.00038
Wickam H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
Acknowledgements
We thank the agricultural technical staff of the JKI, who enabled the laborious growing of the field trials. We thank the team from 3cr Bioscience for the always-fast primer design.
Funding
Open Access funding enabled and organized by Projekt DEAL. This project was funded by the "Fachagentur Nachwachsende Rohstoffe" (FNR) on behalf of the Federal Ministry of Food and Agriculture (BMEL) (Funding ID: 22023215). The funding was supported by Dr. Junghanns GmbH, Aschersleben OT Groß Schierstedt; BBG Kilian-Horsch GbR, Aschersleben OT Mehringen; Horrmann GbR, Bördeland OT Welsleben; Landwirtschaftsbetrieb Hans-Eckhard Kittler, Aschersleben. A fund for open access publication was granted by the Institute of Federal Research Centre for Cultivated Plants, Julius Kuehn-Institute (JKI), Quedlinburg, Germany.
Author information
Authors and Affiliations
Contributions
DvM designed and conducted the trial. JB supported the implementation of the DNA extraction protocol for caraway. DvM performed statistical analyses. DvM investigated literature and wrote the original draft. FM, HL, JB and WJ revised the draft. FM and WJ were responsible for project planning, administration and funding acquisition. All authors read and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
We disclosed all entities, which supported the funding of the project.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Fig. S1
Polycross of three Latin squares to outcross 18 caraway inbred lines. Each Latin square contained 18 plants per line. The trial was grown at the JKI location in Quedlinburg in 2018. By Daniel von Maydell, 2018/07/03 (TIF 7939 kb)
Fig. S2
Minimal and maximal temperature and precipitation at the JKI location in Quedlinburg in 2018 (TIF 1461 kb)
Table S1
Data summary for single lines or F1 populations, respectively, of various traits gathered in a polycross or separate trial in 2018 (XLSX 13 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
von Maydell, D., Brandes, J., Lehnert, H. et al. Breeding synthetic varieties in annual caraway: observations on the outcrossing rate in a polycross using a high-throughput genotyping system. Euphytica 217, 1 (2021). https://doi.org/10.1007/s10681-020-02732-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-020-02732-5