Abstract
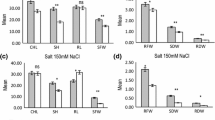
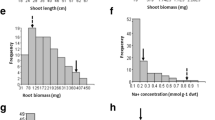
Soil salinity is major constraint for wheat production globally and breeding wheat cultivars for salt tolerance by conventional means is difficult. Therefore, understanding molecular components associated with salt tolerance is needed to facilitate breeding for salt tolerance in wheat. In this investigation, quantitative trait loci (QTL/s) associated with salt tolerance were identified using recombinant inbred lines (RILs) developed from a cross between Kharchia 65 (KH 65) and HD 2009 cultivars. Parents and RILs were evaluated under controlled and sodic stress conditions for 11 morpho-physiological and yield determining traits for two consecutive crop cycles. Simple sequence repeat (SSR) markers were employed for mapping studies. Using composite interval mapping approach, 11 QTLs on 6 chromosomal regions (1B, 2D, 5D, 6A, 6B and 7D) for 7 different traits were identified explaining proportion of the phenotypic variance (PVEs) (2.5–12.8%) under control condition. Three of the QTLs (QCph.iiwbr-2D.1, QCle.iiwbr-6A and QCle.iiwbr-6B) were most consistent in all the environments and explained PVEs (5.1–12.8%) under control condition. Twenty-five QTLs were detected on 7 chromosomal regions (1A, 1B, 2D, 4D, 5D, 6A and 7D) for 10 different traits explaining PVEs (2.6–15.1%) under salt stress. Six of the QTLs namely QSNa+.iiwbr-1B, QSK+.iiwbr-2D, QStn.iiwbr-4D, QSph.iiwbr-2D.1, QSph.iiwbr-6A and QSdth.iiwbr-2D were consistently reproducible in all the environments and explained PVEs ranging from 2.6 to 15.1%. SSR markers namely gwm 261, wmc 112, and cfd 84 were tightly linked with QTLs for K+ content; DTH and DTA; and TN and NE, respectively. Several QTLs contributing towards salt tolerance were present on 2D chromosome. Most of the QTLs linked with salt tolerant traits were inherited from KH 65 signifying the presence of several genes associated with salt tolerance in this cultivar. The information is very useful in marker assisted breeding to enhance salt tolerance in wheat.





Similar content being viewed by others
References
Akbarpour OA, Dehghani H, Rousta MJ (2015) Evaluation of salt stress of Iranian wheat germplasm under field conditions. Crop Pasture Sci 66:770–781
Amin AY, Diab AA (2013) QTL mapping of wheat (Triticum aestivum L.) in responce to salt stress. Int J Bio-Technol Res 3:47–60
Amin A, Eslam MH, Seyed AM et al (2011) Screening of recombinant inbred lines for salinity tolerance in bread wheat (Triticum aestivum L.). Afr J Biotechnol 10:12875–12881. https://doi.org/10.5897/AJB11.1729
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Asif MA, Schilling RK, Tilbrook J et al (2018) Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population. Theor Appl Genet 131:2179–2196. https://doi.org/10.1007/s00122-018-3146-y
Ayed-Slama O, Bouhaouel I, Chamekh Z et al (2018) Genetic variation of salt-stressed durum wheat (Triticum turgidum subsp. durum Desf.) genotypes under field conditions and gynogenetic capacity. J Genet Eng Biotechnol 16:161–167. https://doi.org/10.1016/j.jgeb.2017.11.004
Azadi A, Mardi M, Hervan EM et al (2015) QTL mapping of yield and yield components under normal and salt-stress conditions in bread wheat (Triticum aestivum L.). Plant Mol Biol Rep 33:102–120. https://doi.org/10.1007/s11105-014-0726-0
Bağci SA, Ekiz H, Yilmaz A (2007) Salt tolerance of sixteen wheat genotypes during seedling growth. Turk J Agric For 31:363–372
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Benito C, Figueiras AM, Zaragoza C et al (1993) Rapid identification of Triticeae genotypes from single seeds using the polymerase chain reaction. Plant Mol Biol 21:181–183
Blum A (1988) Plant breeding for stress environments. CRC Press, Boca Raton
Börner A, Schumann E, Fürste A et al (2002) Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor Appl Genet 105:921–936
Byrt CS, Platten JD, Spielmeyer W et al (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1 [OA]. Plant Physiol 143:1918–1928. https://doi.org/10.1104/pp.106.093476
Chen Z, Pottosin II, Cuin TA et al (2007) Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol 145:1714–1725
CSSRI (2015) CSSRI vision 2050. Central soil salinity research institute, Karnal, Haryana, India. http://www.cssri.org. Accessed 28 Oct 2018
Cuin TA, Betts SA, Chalmandrier R, Shabala S (2008) A root’s ability to retain K+ correlates with salt tolerance in wheat. J Exp Bot 59:2697–2706
Cuthbert JL, Somers DJ, Brûlé-Babel AL et al (2008) Molecular mapping of quantitative trait loci for yield and yield components in spring wheat (Triticum aestivum L.). Theor Appl Genet 117:595–608. https://doi.org/10.1007/s00122-008-0804-5
del Pozo A, Yáñez A, Matus IA et al (2016) Physiological traits associated with wheat yield potential and performance under water-stress in a mediterranean environment. Front Plant Sci 7:987
Díaz De León JL, Escoppinichi R, Geraldo N et al (2011) Quantitative trait loci associated with salinity tolerance in field grown bread wheat. Euphytica 181:371–383. https://doi.org/10.1007/s10681-011-0463-5
Dubcovsky J, Maria GS, Epstein E (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet 92:448–454
El-Hendawy SE, Hassan WM, Al-Suhaibani NA et al (2017) Comparative performance of multivariable agro-physiological parameters for detecting salt tolerance of wheat cultivars under simulated saline field growing conditions. Front Plant Sci 8:435
FAO (2014) Land and plant nutrition management service. FAO soils portal. Food and agriculture organization of the united nations. http://www.fao.org. Accessed 28 Oct 2018
Farshadfar E, Mahjouri S, Aghaee M (2008) Detection of epistasis and estimation of additive and dominance components of genetic variation for drought tolerance in durum wheat. J Biol Sci 8:598–603
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319. https://doi.org/10.1093/jxb/erh003
Foolad MR, Jones RA (1993) Mapping salt-tolerance genes in tomato (Lycopersicon esculentum) using trait-based marker analysis. Theor Appl Genet 87:184–192
Fukuda A, Nakamura A, Tanaka Y (1999) Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa 1. Biochim Biophys Acta 1446:149–155
Genc Y, Oldach K, Verbyla AP et al (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor Appl Genet 121:877–894. https://doi.org/10.1007/s00122-010-1357-y
Ghaedrahmati M, Mardi M, Naghavi MR et al (2014) Mapping QTLs associated with salt tolerance related traits in seedling stage of wheat (Triticum aestivum L.). J Agric Sci Technol 16:1413–1428
Gitonga VW, Koning-Boucoiran CF, Verlinden K et al (2014) Genetic variation, heritability and genotype by environment interaction of morphological traits in a tetraploid rose population. BMC Genet 15:146
Heidari B, Sayed-Tabatabaei BE, Saeidi G et al (2011) Mapping QTL for grain yield, yield components, and spike features in a doubled haploid population of bread wheat. Genome 54:517–527. https://doi.org/10.1139/g11-017
Huang XQ, Cöster H, Ganal MW, Röder MS (2003) Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor Appl Genet 106:1379–1389
Huang XQ, Kempf H, Ganal MW, Röder MS (2004) Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theor Appl Genet 109:933–943
Huang XQ, Cloutier S, Lycar L et al (2006) Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor Appl Genet 113:753–766
Hussain B, Lucas SJ, Ozturk L, Budak H (2017) Mapping QTLs conferring salt tolerance and micronutrient concentrations at seedling stage in wheat. Sci Rep 7:15662. https://doi.org/10.1038/s41598-017-15726-6
James RA, Davenport RJ, Munns R (2006) Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2 1. Plant Physiol 142:1537–1547. https://doi.org/10.1104/pp.106.086538
Khan MA, Shirazi MU, Khan MA et al (2009) Role of proline, K/Na ratio and chlorophyll content in salt tolerance of wheat (Triticum aestivum L.). Pak J Bot 41:633–638
Kuchel H, Williams KJ, Langridge P et al (2007) Genetic dissection of grain yield in bread wheat. I. QTL analysis. Theor Appl Genet 115:1029–1041. https://doi.org/10.1007/s00122-007-0629-7
Kumar N, Kulwal PL, Balyan HS, Gupta PK (2007) QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol Breed 19:163–177
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374. https://doi.org/10.1534/genetics.106.066811
Lindsay MP, Lagudah ES, Hare RA, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol 31:1105–1114. https://doi.org/10.1071/FP04111
Liu Y, Liu Y, Zhang Q et al (2018) Genome-wide association analysis of quantitative trait loci for salinity-tolerance related morphological indices in bread wheat. Euphytica 214:176. https://doi.org/10.1007/s10681-018-2265-5
Masoudi B, Mardi M, Hervan EM et al (2015) QTL mapping of salt tolerance traits with different effects at the seedling stage of bread wheat. Plant Mol Biol Rep 33:1790–1803. https://doi.org/10.1007/s11105-015-0874-x
Mba C, Afza R, Jain SM et al (2007) Induced mutations for enhancing salinity tolerance in rice. In: Jenks MA, Hasegawa PM, Jain SM (eds) Advances in molecular breeding toward drought and salt tolerant crops. Springer, Berlin, pp 413–454
Mbarki S, Sytar O, Zivcak M et al (2018) Anthocyanins of coloured wheat genotypes in specific response to salt stress. Molecules 23:1518
Mclntosh RA (1998) Catalogue of gene symbols for wheat. In: 9th international wheat genetics symposium. Saskatoon, Canada, pp 123–142
Mujeeb-Kazi A, Munns R, Rasheed A et al (2019) Breeding strategies for structuring salinity tolerance in wheat. In: Sparks DL (ed) Advances in agronomy. Elsevier, Amsterdam, pp 121–187
Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253:201–218
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Munns R, Wallace PA, Teakle NL, Colmer TD (2010) Measuring soluble ion concentrations (Na+, K+, Cl-) in salt-treated plants. In: Sunkar R (ed) Plant stress tolerance. Springer, Berlin, pp 371–382
Oyiga BC, Sharma RC, Shen J et al (2016) Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J Agron Crop Sci 202:472–485. https://doi.org/10.1111/jac.12178
Oyiga BC, Sharma RC, Baum M et al (2018) Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat: GWAS reveals gene loci for salt tolerance in wheat. Plant, Cell Environ 41:919–935. https://doi.org/10.1111/pce.12898
Paliwal R, Röder MS, Kumar U et al (2012) QTL mapping of terminal heat tolerance in hexaploid wheat (T. aestivum L.). Theor Appl Genet 125:561–575. https://doi.org/10.1007/s00122-012-1853-3
Poustini K, Siosemardeh A (2004) Ion distribution in wheat cultivars in response to salinity stress. Field Crops Res 85:125–133
Rana V, Ram S, Sedhil R et al (2015) Physiological, biochemical and morphological study in wheat (Triticum aestivum L.) RILs population for salinity tolerance. J Agric Sci 7:119–128
Rana V, Ram S, Nehra K, Sharma I (2016) Expression of genes related to Na+ exclusion and proline accumulation in tolerant and susceptible wheat genotypes under salt stress. Cereal Res Commun 44:404–413. https://doi.org/10.1556/0806.44.2016.009
Ren Z-H, Gao J-P, Li L-G et al (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146
Ren Z, Zheng Z, Chinnusamy V et al (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci 107:1–6
Rengasamy P (2010) Soil processes affecting crop production in salt-affected soils. Funct Plant Biol 37:613. https://doi.org/10.1071/FP09249Ren
Reynolds MP, Balota M, Delgado MIB et al (1994) Physiological and morphological traits associated with spring wheat yield under hot, irrigated conditions. Funct Plant Biol 21:717–730
Ribaut J-M, Jiang C, Gonzalez-de-Leon D et al (1997) Identification of quantitative trait loci under drought conditions in tropical maize. 2. Yield components and marker-assisted selection strategies. Theor Appl Genet 94:887–896
Roder MS, Korzun V, Wendehake K et al (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Sairam RK, Srivastava GC, Agarwal S, Meena RC (2005) Differences in antioxidant activity in response to salinity stress in tolerant and susceptible wheat genotypes. Biol Plant 49:85–91
Shamaya NJ, Shavrukov Y, Langridge P et al (2017) Genetics of Na+ exclusion and salinity tolerance in Afghani durum wheat landraces. BMC Plant Biol 17:209. https://doi.org/10.1186/s12870-017-1164-6
Sharma R, Rao B, Saxena R (2004) Salt affected soils in India—current assessment. In: Advances in sodic land reclamation. 9–14 February, Lucknow, India, pp 1–26
Siddiqui MN, Mostofa MG, Akter MM et al (2017) Impact of salt-induced toxicity on growth and yield-potential of local wheat cultivars: oxidative stress and ion toxicity are among the major determinants of salt-tolerant capacity. Chemosphere 187:385–394
Snedecor GW, Cochran WG (1980) Statistical methods, 7th edn. Iowa State University Press, Ames, Iowa, USA
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114. https://doi.org/10.1007/s00122-004-1740-7
Spielmeyer W, Hyles J, Joaquim P et al (2007) A QTL on chromosome 6A in bread wheat (Triticum aestivum) is associated with longer coleoptiles, greater seedling vigour and final plant height. Theor Appl Genet 115:59–66. https://doi.org/10.1007/s00122-007-0540-2
Srinivasa J, Arun B, Mishra VK et al (2014) Zinc and iron concentration QTL mapped in a Triticum spelta × T. aestivum cross. Theor Appl Genet 127:1643–1651. https://doi.org/10.1007/s00122-014-2327-6
Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97. https://doi.org/10.1016/j.tplants.2009.11.009
Tavakkoli E, Fatehi F, Coventry S et al (2011) Additive effects of Na+ and Cl– ions on barley growth under salinity stress. J Exp Bot 62:2189–2203
Tiwari C, Wallwork H, Kumar U et al (2013) Molecular mapping of high temperature tolerance in bread wheat adapted to the Eastern Gangetic Plain region of India. Field Crops Res 154:201–210. https://doi.org/10.1016/j.fcr.2013.08.004
Tuberosa R, Sanguineti MC, Landi P et al (2002) Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol Biol 48:697–712
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Xu YF, An DG, Liu DC et al (2012) Mapping QTLs with epistatic effects and QTL × treatment interactions for salt tolerance at seedling stage of wheat. Euphytica 186:233–245. https://doi.org/10.1007/s10681-012-0647-7
Xu Y, Li S, Li L et al (2013) Mapping QTLs for salt tolerance with additive, epistatic and QTL × treatment interaction effects at seedling stage in wheat. Plant Breed 132:276–283. https://doi.org/10.1111/pbr.12048
Yeo AR, Yeo ME, Flowers SA, Flowers TJ (1990) Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor Appl Genet 79:377–384
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421. https://doi.org/10.1111/j.1365-3180.1974.tb01084.x
Zhang LP, Xu XQ, Zhao CP et al (2011) QTL analysis of plant height in photoperiod-thermo sensitive male sterile wheat. Mol Plant Breed 2:92–97
Zhu J-K, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10:1181–1191
Acknowledgements
The authors would like to extend their sincere appreciation to the support and facilities provided by ICAR-Indian Institute of Wheat and Barley Research (IIWBR), Karnal, India under the network project for salt tolerance funded by Indian Council of Agricultural Research (ICAR), New Delhi, India. We are grateful for the support of RD for conducting experiment, data collection, analysis and manuscript writing, SR for overall guidance in conducting experiments, analysis of data and writing manuscript, VR for generating data on SSR, VKM for data analysis and manuscript writing, VP and GPS for reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Devi, R., Ram, S., Rana, V. et al. QTL mapping for salt tolerance associated traits in wheat (Triticum aestivum L.). Euphytica 215, 210 (2019). https://doi.org/10.1007/s10681-019-2533-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-019-2533-z