Abstract
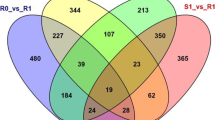
The oomycete Plasmopara viticola (Berk. & Curt.) Berl. & de Toni causes downy mildew, one of the most devastating diseases of grapevine (Vitis vinifera L.). Traditional European grapevine cultivars are highly susceptible to this obligate biotrophic pathogen. Large amounts of fungicides are necessary to protect the grapevine plants and secure harvest. This strong requirement for protective chemicals conflicts with the modern demand for sustainability in agriculture. A significant reduction of chemical protection is possible by generating novel robust grapevine cultivars through resistance breeding. Current grapevine breeding utilizes marker-assisted genetic selection. The aim is to combine diverse resistance loci for durable resistance. Markers tagging various resistance loci were elaborated during the last 10 years. However, knowledge about the conveyed defense mechanisms is still sparse but would be essential to optimize the combination of resistance loci. Asian Vitis amurensis accessions carry resistance against downy mildew e.g. in the Rpv10 locus. This locus has been introgressed into the resistant grapevine cultivar ‘Solaris’ and was genetically mapped to chromosome nine. To understand its mode of action in early defense reactions we performed a comparative RNA sequencing analysis after pathogen challenge of Rpv10-carriers, Rpv10-carriers containing additionally the resistance locus Rpv3 from American Vitis sp. origin and non-Rpv carriers. This study indicated comprehensive transcriptional re-programming and a large number of differentially expressed genes. The data indicates that the difference between resistant and susceptible grapevines relies in the increased amount of responsive genes and the efficiency of early signal transduction. This results in the fast activation of large gene clusters encoding phenylalanine ammonium lyase and stilbene synthase on chromosome 16.




Similar content being viewed by others
References
Ali K, Maltese F, Figueiredo A, Rex M, Fortes AM, Zyprian E, Pais MS, Verpoorte R, Choi YH (2012) Alterations in grapevine leaf metabolism upon inoculation with Plasmopara viticola in different time-points. Plant Sci 191:100–107. https://doi.org/10.1016/j.plantsci.2012.04.014
Alonso-Villaverde V, Voinesco F, Viret O, Spring JL, Gindro K (2011) The effectiveness of stilbenes in resistant Vitaceae: ultrastructural and biochemical events during Plasmopara viticola infection process. Plant Physiol Biochem 49(3):265–274. https://doi.org/10.1016/j.plaphy.2010.12.010
Bellin D, Peressotti E, Merdinoglu D, Wiedemann-Merdinoglu S, Adam-Blondon AF, Cipriani G, Morgante M, Testolin R, Di Gaspero G (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genet 120(1):163–176. https://doi.org/10.1007/s00122-009-1167-2
Canaguier A, Grimplet J, Di Gaspero G, Scalabrin S, Duchêne E, Choisne N, Mohellibi N, Guichard C, Rombauts S, Le Clainche I, Bérard A, Chauveau A, Bounon R, Rustenholz C, Morgante M, Le Paslier M-C, Brunel D, Adam-Blondon A-F (2017) A new version of the grapevine reference genome assembly (12X. v2) and of its annotation (VCost. v3). Genom Data 14:56
Casagrande K, Falginella L, Castellarin SD, Testolin R, Di Gaspero G (2011) Defence responses in Rpv3-dependent resistance to grapevine downy mildew. Planta 234(6):1097–1109. https://doi.org/10.1007/s00425-011-1461-5
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124(4):803–814. https://doi.org/10.1016/j.cell.2006.02.008
Chong JL, Poutaraud A, Hugueney P (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177(3):143–155. https://doi.org/10.1016/j.plantsci.2009.05.012
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138(3):963–971
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411(6839):826–833. https://doi.org/10.1038/35081161
Delmotte F, Mestre P, Schneider C, Kassemeyer HH, Kozma P, Richart-Cervera S, Rouxel M, Deliere L (2013) Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect Genet Evolut. https://doi.org/10.1016/j.meegid.2013.10.017
Di Gaspero G, Copetti D, Coleman C, Castellarin SD, Eibach R, Kozma P, Lacombe T, Gambetta G, Zvyagin A, Cindric P, Kovacs L, Morgante M, Testolin R (2012) Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor Appl Genet 124(2):277–286. https://doi.org/10.1007/s00122-011-1703-8
Dudenhöffer J, Schwander F, Töpfer R, Zyprian E (2015) Sequence analysis of loci Rpv10 and Rpv3 for resistance against grapevine downy mildew (Plasmopara viticola). In: Shao-Hua L et al (eds) ISHS Proceedings of XIth international conference on grapevine breeding and genetics. Acta Hort 1082:69–72
Eibach R, Zyprian E, Welter L, Töpfer R (2007) The use of molecular markers for pyramiding resistance genes in grapevine breeding. Vitis 46(3):120–124
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM, Grando MS (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13:39
Eurostat (2007) The use of plant protection products in the European Union (1992–2003). http://ec.europa.eu/eurostat/de/data/database. Accessed 10 Jan 2019
Fernandes H, Michalska K, Sikorski M, Jaskolski M (2013) Structural and functional aspects of PR-10 proteins. FEBS J 280:1169–1199
Figueiredo A, Fortes AM, Ferreira S, Sebastiana M, Choi YH, Sousa L, Acioli-Santos B, Pessoa F, Verpoorte R, Pais MS (2008) Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J Exp Bot 59(12):3371–3381. https://doi.org/10.1093/jxb/ern187
Figueiredo A, Monteiro F, Fortes AM, Bonow-Rex M, Zyprian E, Sousa L, Pais MS (2012) Cultivar-specific kinetics of gene induction during downy mildew early infection in grapevine. Funct Integr Genom 12(2):379–386. https://doi.org/10.1007/s10142-012-0261-8
Figueiredo J, Costa GJ, Maia M, Paulo OS, Malho R, Silva MS, Figueiredo A (2016) Revisiting Vitis vinifera subtilase gene family: a possible role in grapevine resistance against Plasmopara viticola. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01783
Fischer BM, Salakhutdinov I, Akkurt M, Eibach R, Edwards KJ, Töpfer R, Zyprian EM (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet 108(3):501–515. https://doi.org/10.1007/s00122-003-1445-3
Fry WE, Grünwald NJ (2010) Introduction to oomycetes. Plant Health Instr. https://doi.org/10.1094/phi-i-2010-1207-01
Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW (2012) Plant disease resistance genes: current status and future directions. Physiol Mol Plant Pathol 78:51–65. https://doi.org/10.1016/j.pmpp.2012.01.002
Hammond-Kosack KE, Jones JD (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607. https://doi.org/10.1146/annurev.arplant.48.1.575
Hood ME, Shew HD (1996) Applications of KOH-aniline blue fluorescence in the study of plant-fungal interactions. Phytopathology 86(7):704–708
Jain S, Kumar A (2015) The pathogenesis related class 10 proteins in plant defense against biotic and abiotic stresses. Adv Plants Agric Res 3(1):00077. https://doi.org/10.15406/apar.2015.03.00077
Jones JD (2001) Putting knowledge of plant disease resistance genes to work. Curr Opin Plant Biol 4(4):281–287
Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329. https://doi.org/10.1038/nature05286
Kortekamp A, Zyprian E (1999) Leaf hairs as a basic protective barrier against downy mildew of grape. J Phytopathol 147(7–8):453–459. https://doi.org/10.1111/j.1439-0434.1999.tb03850.x
Kortekamp A, Zyprian E (2003) Characterization of Plasmopara-resistance in grapevine using in vitro plants. J Plant Physiol 160(11):1393–1400. https://doi.org/10.1078/0176-1617-01021
Kortekamp A, Wind R, Zyprian E (1998) Investigation of the interaction of Plasmopara viticola with susceptible and resistant grapevine cultivars. Z Pflanzenk Pflanzen 105(5):475–488
Kortekamp A, Wind R, Zyprian E (1999) The role of hairs on the wettability of grapevine (Vitis spp.) leaves. Vitis 38(3):101–105
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121(1):185–199
Liu JJ, Ekramoddoullah AKM, Yu X (2003) Differential expression of multiple PR10 proteins in western white pine following wounding, fungal infection and cold-hardening. Physiol Plant 119:544–553
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Lloyd S (1982) Least squares quantization in PCM. IEEE Trans Inf Theor 28(2):129–137. https://doi.org/10.1109/tit.1982.1056489
Malacarne G, Vrhovsek U, Zulini L, Cestaro A, Stefanini M, Mattivi F, Delledonne M, Velasco R, Moser C (2011) Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol. https://doi.org/10.1186/1471-2229-11-114
Malinovsky FG, Fangel JU, Willats WG (2014) The role of the cell wall in plant immunity. Front Plant Sci 5:178. https://doi.org/10.3389/fpls.2014.00178
Mestre P, Carrere S, Gouzy J, Piron MC, de Labrouhe DT, Vincourt P, Delmotte F, Godiard L (2016) Comparative analysis of expressed CRN and RXLR effectors from two Plasmopara species causing grapevine and sunflower downy mildew. Plant Pathol 65(5):767–781. https://doi.org/10.1111/ppa.12469
Mohr HD (2011) Farbatlas Krankheiten, Schädlinge und Nützlinge an der Weinrebe, 2nd edn. Eugen Ulmer, Stuttgart
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628. https://doi.org/10.1038/nmeth.1226
Moser T (2015) Untersuchung der transkriptionellen Regulation von Kandidatengenen der Pathogenabwehr gegen Plasmopara viticola in der Weinrebe. http://dx.doi.org/10.5073/dissjki.2015.004
Nicholas P, Magarey P, Wachtel M (1994) Diseases and pests: grape production series no. 1. Winetitles. Adelaide, Australia
Niderman T, Genetet I, Bruyère T, Gees R, Stintzi A, Legrand M, Fritig B, Mösinger E (1995) Pathogenesis-Related PR-1 proteins are antifungal—isolation and characterization of three 14-kilodalton proteins of tomato and of a Basic PR-1 of Tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol 108:17–27
Nürnberger T, Lipka V (2005) Non-host resistance in plants: new insights into an old phenomenon. Mol Plant Pathol 6(3):335–345. https://doi.org/10.1111/j.1364-3703.2005.00279.x
Oliveros JC (2007–2015) An interactive tool for comparing lists with Venn’s diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html. Accessed 10 Jan 2019
Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139(3):1185–1193. https://doi.org/10.1104/pp.105.066324
Parage C, Tavares R, Réty S, Baltenweck-Guyot R, Poutaraud A, Renault L, Heintz D, Lugan R, Marais G, Aubourg S, Hugueney P (2012). Structural, functional and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine (Vitis vinifera). Plant Phys 160(3):1407–1419. https://doi.org/10.1105/pp.112.202705
Park MR, Yun KY, Mohanty B, Herath V, Xu F, Wijaya E, Bajic VB, Yun SJ, De Los Reyes BG (2010) Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ 33(12):2209–2230. https://doi.org/10.1111/j.1365-3040.2010.02221.x
Peressotti E, Wiedemann-Merdinoglu S, Delmotte F, Bellin D, Di Gaspero G, Testolin R, Merdinoglu D, Mestre P (2010) Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol 10:147. https://doi.org/10.1186/1471-2229-10-147
Pinto PM, Ricardo PPC (1995) Lupinus albus L. pathogenesis-related proteins that show similarity to PR-10 proteins. Plant Physiol 109:1345–1351
Polesani M, Bortesi L, Ferrarini A, Zamboni A, Fasoli M, Zadra C, Lovato A, Pezzotti M, Delledonne M, Polverari A (2010) General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genom. https://doi.org/10.1186/1471-2164-11-117
Salmaso M, Faes G, Segala C, Stefanini M, Salakhutdinov I, Zyprian E, Toefer R, Grando MS, Velasco R (2004) Genome diversity and gene haplotypes in the grapevine (Vitis vinifera L.), as revealed by single nucleotide polymorphisms. Mol Breed 14(4):385–395. https://doi.org/10.1007/s11032-004-0261-z
Santamaria ME, Martinez M, Cambra I, Grbic V, Diaz I (2013) Understanding plant defence responses against herbivore attacks: an essential first step towards the development of sustainable resistance against pests. Transgenic Res 22(4):697–708. https://doi.org/10.1007/s11248-013-9725-4
Schwander F, Eibach R, Fechter I, Hausmann L, Zyprian E, Töpfer R (2012) Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor Appl Genet 124(1):163–176. https://doi.org/10.1007/s00122-011-1695-4
Sekhwal MK, Li P, Lam I, Wang X, Cloutier S, You FM (2015) Disease resistance gene analogs (RGAs) in plants. Int J Mol Sci 16(8):19248–19290. https://doi.org/10.3390/ijms160819248
Sels J, Mathys J, De Coninck BMA, Cammue BPA, Bolle MFC (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46(11):941–950. https://doi.org/10.1016/j.plaphy.2008.06.011
Somssich EI, Schmelzer E, Kawalleck P, Hahlbrock K (1988) Gene structure and in situ transcript localization of pathogenesis-related protein 1 in parsley. Mol Gen Genet 213:93–98
Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ (2002) The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14(11):2929–2939
Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJ, Takken FL (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140(4):1233–1245. https://doi.org/10.1104/pp.105.073510
Töpfer R, Hausmann L, Eibach R (2011) Molecular Breeding. In: Adam-Blondon AF, Martinez-Zapater JM, Kole C (eds) Genetics, genomics and breeding of grapes. Science Publishers, Enfield
Van der Biezen EA, Jones JD (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23(12):454–456
Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Ann Rev Phytopathol 44:135–162. https://doi.org/10.1146/annurev.phyto.44.070505.143425
Welter LJ, Göktürk-Baydar N, Akkurt M, Maul E, Eibach R, Töpfer R, Zyprian EM (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol Breed 20(4):359–374. https://doi.org/10.1007/s11032-007-9097-7
Welter L, Tisch C, Kortekamp A, Töpfer R, Zyprian E (2017) Powdery mildew responsive genes of resistant grapevine cultivar ‘Regent’. Vitis 56:181–188
Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PRJ (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450(7166):115–118. http://www.nature.com/nature/journal/v450/n7166/suppinfo/nature06203_S1.html
Yin L, An Y, Qu J, Li X, Zhang Y, Dry I, Wu H, Lu J (2017) Genome sequence of Plasmopara viticola and insight into the pathogenic mechanism. Sci Rep 7:46553. https://doi.org/10.1038/srep46553
Acknowledgements
This project was co-financed by the European Union—European Regional Development Fund (ERDF) (A23 partial)—as part of the program INTERREG IV Upper Rhine (“Transcending borders with every project”). It was additionally supported by the Research Foundation of German Viticulture of the German Association of Agriculture (Forschungsring des Deutschen Weinbaus bei der DLG) (Rpv10). We wish to thank the Breeding Department of the Institute of Grapevine Breeding Geilweilerhof for the contribution of data on markers linked to Plasmopara viticola resistance in grapevine genetic resources.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fröbel, S., Dudenhöffer, J., Töpfer, R. et al. Transcriptome analysis of early downy mildew (Plasmopara viticola) defense in grapevines carrying the Asian resistance locus Rpv10. Euphytica 215, 28 (2019). https://doi.org/10.1007/s10681-019-2355-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-019-2355-z