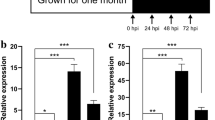
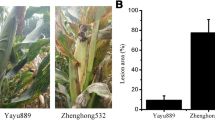
Abstract
Common leaf spot (CLS) which caused by Pseudopeziza medicaginis is a serious foliar disease in alfalfa. Here, we report a differential expression model of transcriptome between leaves from a CLS-resistant line (RI vs. Rck) and a CLS-susceptible line (SI vs. Sck) after inoculation with P. medicaginis using RNA-seq technology on genome-wide. De novo assembling of clean reads generated 83,625 transcripts with 35,061 unigenes, of which, 85% were found to be able to match the reference genes. 3304 differentially expressed genes (DEGs) were detected in RI versus Rck library while 4276 DEGs were found in SI versus Sck library. Gene ontology function analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of these DEGs were performed in this work, and the results showed that some categories were significantly enriched in RI versus Rck library and not in SI versus Sck library including response to stimulus, response to stress, oxidation–reduction process, and oxidoreductase activity while KEGG pathway analysis suggested that plant-pathogen interaction was one of the main biological pathways in which DEGs in RI versus Rck library were enriched, but not that for DEGs in SI versus Sck library. Furthermore, about only 20 DEGs showed obviously being up-regulated in RI versus RCK library and being down-regulated or not being detected in SI versus SCK library and the analysis on the signal transduction pathways related to these DEGs indicated that both SA-mediated signal transduction pathway and ET-mediated signal transduction pathway were related to the CLS resistance of alfalfa. In this work, we comprehensively and systematically characterized the molecular basis of alfalfa in response to P. medicaginis for the first time. The information resulted from this study is useful for future investigating the mechanism of CLS resistance on molecular level in alfalfa.







Similar content being viewed by others
References
Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63:3523–3543
Barnes DK, Goplen RP, Baylor JE (1988) Alfalfa and alfalfa improvement. American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America, Madison
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284
Chen W, Zou G, Whitham SA, Budworth PR, Tao Y, Xie Z et al (2002) Expression profile matrix of Arabidopsis transcription factor genes predict their putative functions in response to environmental stresses. Plant Cell 14(3):559–574
Chung HS, Howe GA (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21(1):131–145
Conesa A, Göz S, García-Gómez JM, Terol J, Talón M, Robles M et al (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
dos Reis SP, Lima AM, de Souza CRB (2012) Recent molecular advances on downstream plant responses to abiotic stress. Int J Mol Sci 13:8628–8647
Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JD (2000) cDNA-AFLP reveals a striking overlap in the race-specific resistance and wound response gene expression profiles. Plant Cell 12(6):963–977
Fan H, Wang F, Gao H, Wang L, Xu J, Zhao Z (2011) Pathogen-induced mdWRKY1 in ‘Qinguan’ apple enhances disease resistance. J Plant Biol 54(3):150–158
Fan XD, Wang JQ, Yang N, Dong YY, Liu L, Wang FW et al (2013) Gene expression profiling of soybean leaves and roots under salt, saline-alkali and drought stress by high-throughput Illumina sequencing. Gene 512:392–402
Gao J, Chen Z, Luo M, Peng H, Lin H, Qin C et al (2014) Genome expression profile analysis of the maize sheath in response to inoculation to R. solani. Mol Biol Rep 41:2471–2483
He R, Kim MJ, Nelson W, Balbuena TS, Kim R, Kramer R et al (2012) Next-generation sequencing-based transcriptomic and proteomic analysis of the common reed, Phragmites australis (Poaceae), reveals genes involved in invasiveness and rhizome specificity. Am J Bot 99(2):232–247. https://doi.org/10.3732/ajb.1100429
Kakumanu A, Ambavaram MMR, Klumas C, Krishnan A, Batlang U, Myers E et al (2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-seq. Plant Physiol 160(2):846
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNASeq data with or without a reference genome. BMC Bioinform 12:323
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16(2):319–331
Li X, Acharya A, Farmer AD, Crow JA, Bharti AK, Kramer RS et al (2012) Prevalence of single nucleotide polymorphism among 27 diverse alfalfa genotypes as assessed by transcriptome sequencing. BMC Genomics 13:568
Liu M, Gao J, Yin F, Gong G, Qin C, Ye K et al (2015) Transcriptome analysis of maize leaf systemic symptom infected by Bipolaris zeicola. PLoS ONE 10(3):e0119858. https://doi.org/10.1371/journal.pone.0119858
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 25:402–408
Mainer A, Leath KT (1978) Foliar diseases alter carbohydrate and protein levels in leaves of alfalfa and orchardgrass. Phytopathology 68:1252–1255
Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA et al (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26(4):403–410
Meng F, Yuan QH, Su DR, Gao JM (2008) Selection and validation of ISSR markers of common leaf spot disease resistance-related gene in tetraploid alfalfa. Plant Prot 34(4):46–50 (in Chinese)
Morgan WC, Parbery DG (1977) Effects of Pseudopeziza leaf spot disease on growth and yield in Lucerne. Aust J Agric Res 28:1029–1040
Morgan WC, Parbery DG (1980) Depressed fodder quality and increased oestrogenic activity of lucerne infected with Pseudopeziza medicaginis. Aust J Agric Res 31:1103–1110
Mysore KS, Crasta OR, Tuori RP, Folferts O, Swirshy PB, Martin GB (2002) Comprehensive transcript profiling of the Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32(3):299–315
Mzid R, Marchive C, Blancard D, Deluc L, Barrieu F, Corio-Costet MF et al (2007) Overexpression of VvWRKY2 in tobacco enhances broad resistance to necrotrophic fungal pathogens. Physiol Plant 131(3):434–447
Nutter FW Jr, Guan J, Gotlieb AR, Rhodes LH, Grau CR, Sulc RM (2002) Quantifying alfalfa yield losses caused by foliar diseases in Iowa, Ohio, Wisconsin, and Vermont. Plant Dis 86:269–277
Obert DE, Skinner DZ, Stuteville DL (2000) Association of AFLP markers with downy mildew resistance in autotetraploid alfalfa. Mol Breed 6:287–294
Ozeretskovskaya OL (2007) Immune response. In: Dzhavakhiya VG, Ozeretskovskaya OL, Zinovyeva SV (eds) Comprehensive and molecualr phytopathology. Elsevier, Netherlands, pp 265–314
Pandini A, Fornili A, Fraternali F, Kleinjung J (2013) GSATools: analysis of allosteric communication and functional local motions using a structural alphabet. Bioinformatics 29(16):2053–2055
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S et al (2003) TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19(5):651–652
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15(5):247–258
Sharma J, Chakraverty N (2013) Mechanism of plant tolerance in response to heavy metals. In: Rout GR, Das AB (eds) Molecular stress physiology of plants. Springer India, New Delhi, pp 289–308
Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR et al (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468(7322):400–405. https://doi.org/10.1038/nature09430
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12(6):239–244
Wang W, Wei Z, Lam TW, Wang J (2011) Next generation sequencing has lower sequence coverage and poorer SNP-detection capability in the regulatory regions. Sci Rep 1:55
Wang Y, Bi B, Yuan QH, Li XL, Gao JM (2012a) Association of AFLP and SCAR markers with common leaf spot resistance in autotetraploid alfalfa (Medicago sativa). Genet Mol Res 11(1):606–616
Wang XS, Liu Y, Jia YY, Gu HY, Ma HY, Yu T et al (2012b) Transcriptional responses to drought stress in root and leaf of chickpea seedling. Mol Biol Rep 39:8147–8158
Wang Y, Xu L, Chen Y, Shen H, Gong Y, Limera C et al (2013) Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (Pb) stress with next generation sequencing. PLoS ONE 8(6):e66539. https://doi.org/10.1371/journal.pone.0066539
Ward JA, Ponnala L, Weber CA (2012) Strategies for transcriptome analysis in nonmodel plants. Am J Bot 99:267–276
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S et al (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322
Yan X, Dong C, Yu J, Liu W, Jiang C, Liu J et al (2013) Transcriptome profile analysis of young floral buds of fertile and sterile plants from the self-pollinated offspring of the hybrid between novel restorer line NR1 and Nsa CMS line in Brassica napus. BMC Genomics 14:26. https://doi.org/10.1186/1471-2164-14-29
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z et al (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:293–297
Ye HY, Du H, Tang N, Li XH, Xiong LZ (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71(3):291–305
Zhang S, Song G, Gao J, Li Y, Guo D, Fan Q et al (2014) Transcriptome characterization and differential expression analysis of cold-responsive genes in young spikes of common wheat. J Biotechnol 189:48–57
Zhao Y, Yao B, Zhang M, Wang S, Zhang H, Xiao W et al (2013) Comparative analysis of differentially expressed genes in Sika deer antler at different stages. Mol Biol Rep 40(2):1665–1676
Zheng WJ, Ma L, Zhao JM, Li ZQ, Sun FY, Lu XC (2013) Comparative transcriptome analysis of two rice varieties in response to rice stripe virus and small brown plant hoppers during early interaction. PLoS ONE 8(12):e82126
Acknowledgements
We thank Dr. Yuehui Chao, Beijing Forest University, for his enlightening discussions, and Professor Zhi Gui, Tianjin Agricultural University, for helpful comments on this manuscript.
Funding was provided by the Conservation of Forage Genetic Resources Program (Grant No. 2130135).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10681_2018_2204_MOESM1_ESM.zip
Table S1. GO function annotation of assembled transcripts. Table S2. Up or down regulated genes in RI and RCK libraries. Table S3. Up or down regulated isoforms in RI and RCK libraries. Table S4. Up or down regulated genes in SI and SCK libraries. Table S5. Up or down regulated isoforms in SI and SCK libraries. Table S6. GO function enrichment analysis of up regulated transcripts in RI and RCK libraries. Table S7. GO function enrichment analysis of down regulated transcripts in RI and RCK libraries. Table S8. GO function enrichment analysis of up regulated transcripts in SI and SCK libraries. Table S9. GO function enrichment analysis of down regulated transcripts in SI and SCK libraries. Table S10. KEGG pathway enrichment analysis of up regulated transcripts in RI and RCK libraries. Table S11. KEGG pathway enrichment analysis of down regulated transcripts in RI and RCK libraries. Table S12. KEGG pathway enrichment analysis of up regulated transcripts in SI and SCK libraries. Table S13. KEGG pathway enrichment analysis of down regulated transcripts in SI and SCK libraries. Table S14. DEGs involved in disease resistance in RI and RCK libraries. Table S15. DEGs involved in disease resistance in SI and SCK libraries. (ZIP 26021 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Huang, H., Wang, Y. et al. Transcriptome characterization and differential expression analysis of disease-responsive genes in alfalfa leaves infected by Pseudopeziza medicaginis. Euphytica 214, 126 (2018). https://doi.org/10.1007/s10681-018-2204-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-018-2204-5