Abstract
To make advances in rice molecular design breeding, it is important to understand the genetic background and phenotypes of the parental lines, to known superior allele of the breeding material. In this study, 34 pairs of insertion-deletion (InDel) markers were used to analyze the differentiation of indica–japonica properties in 277 primal parental lines of hybrid rice that are used in China and abroad. The genetic diversity and population structure were assessed with simple sequence repeat (SSR) markers. We also evaluated the association between SSR markers and seven yield traits and four grain-quality traits of 256 indica rice lines. The main results are as follows. (1) Within the 277 lines there were two subgroups: indica and japonica. The 256 lines of the indica-subgroup were further divided into four sub–sub-groups: landrace, maintainer, restorer, and rice lines from the International Rice Research Institute. (2) We identified 29 loci associated with 11 agronomic traits, including one for plant height, two for tiller number, one for panicle length, three for grain length, eight for grain width, eight for grain length/width ratio, one for thousand grain weight, one for amylose content, two for gel consistency, one for gelatinization temperature and one for total protein content. (3) Among those loci, 16 markers were closely linked to previously identified genes. Superior allele carrier lines and allele effects were also analyzed. Our results provided the population structure and indica–japonica properties for parent matching, and the marker–trait association will also be useful for molecular design breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is one of the most important cereal crops in the world, and China is a large rice producer and consumer. Therefore, high and stable rice yields are important for the security of China’s food supply. Moreover, the demand for high quality rice is continuously increasing as living standards improve. Hence, there is a need to breed high quality rice with large yields. Rice yield and grain quality are quantitative traits controlled by multiple minor-effect genes. Identifying these alleles in various lines and elucidating their functions will be important in hybrid rice breeding programs.
Yield-related traits are quantitative traits controlled by multiple minor-effect genes/loci (quantitative trait loci or QTLs) distributed in the 12 rice chromosomes (www.gramene.org). Many yield-related genes have been cloned, such as GS3, GW2, GW5, GL7, Ghd7, OsSPL16, TGW6, GIF1, DTH8 and Gnla (Ashikari et al. 2005; Ishimaru et al. 2013; Mao et al. 2010; Song et al. 2007; Wang et al. 2008, 2012, 2015; Wei et al. 2010; Xue et al. 2008; Yan et al. 2011). Some of these genes have pleiotropic effects: for example, GS3 controls both grain length and weight; Ghd7 controls grain number per spike, plant height, and heading date; GW2 controls grain width, grain weight, panicle number per plant, and heading date; and GL7 controls grain length and width.
Grain quality traits, including rice milling quality, appearance quality, cooking and eating quality, and nutritional quality, are also quantitative traits. Few QTLs for milling quality have been identified. Appearance quality is also associated with yield-related traits, and many QTLs for grain appearance have been identified (Mao et al. 2010; Song et al. 2007; Wang et al. 2008, 2015; Yan et al. 2011). Amylose content (AC), gelatinization temperature (GT), and gel consistency (GC) are the three main properties related to rice cooking and eating quality. Many studies focused on the starch synthesis pathway and 11 genes have been cloned (http://www.ricedata.cn/gene/index.htm). AC is mainly controlled by Wx, SSIIa, and SSIII (Fujita et al. 2007; Sato et al. 2002; Zhang et al. 2011a). Wx is a pleiotropic gene, which also regulates GC. Map-based cloning has confirmed that Wx is the major QTL qGC-6 for GC of japonica/indica cross (Su et al. 2011). More than 20 QTLs for GC have been identified, but only Wx has been cloned. GT is mainly controlled by ALK/SSIIa (Gao et al. 2003). The total protein content (TPC) is the second largest component by weight in the endosperm, but rice lines with protein contents >9 % in the endosperm have poor eating quality. None of the QTLs for TPC distributed throughout all 12 chromosomes have yet been cloned.
As rice breeding has become increasingly difficult, molecular design breeding has become more popular. Although many genes for rice yield and grain quality have been fine-mapped and cloned, more information is required for rice molecular design breeding. First, genes related to rice yield and grain quality are quantitative traits, and various alleles in the different lines cause different phenotypes, but little information is published. Second, the genetic background and phenotypes of parental lines, especially the primal parental lines, are not well known. However, understanding indica–japonica properties is important in parent matching for super hybrid breeding practices. Therefore, we need to understand the various alleles in the germplasm first, and then identify the effect of different alleles and determine the most important alleles controlling target traits.
Genome wide association analysis had been widely conducted on rice to detect complex trait variation (Chen et al. 2014; Huang et al. 2011; Zhao et al. 2011). In this study, we used 277 primal parental lines of hybrid rice and intermediate materials from more than ten provinces in China and Southeast Asian countries that are major rice producing areas. Thirty-four insertion-deletion (InDel) markers were used to analyze the differentiation of indica–japonica properties, and 193 simple sequence repeats (SSR) markers were used to assess the genetic diversity and a SSR subset was applied to population structure. Moreover, association analysis was undertaken between SSR markers, seven yield traits and four grain quality traits of 256 indica rice lines. Our goals were to (1) clarify the indica–japonica properties and distinguish the population structure, (2) identify SSR markers associated with rice yield and grain quality-related traits, and (3) establish allele effects on phenotypes to lay the foundation for parent matching and molecular design breeding.
Materials and methods
Plant materials
A total of 277 hybrid rice primal parental lines and intermediate materials were collected from more than ten rice producing provinces in China, Pakistan, Indonesia, Vietnam and other countries (Table S1) as well as the International Rice Research Institute (IRRI, the Philippines).
Field experiments and phenotypes
During the 2013 and 2014 growing seasons, the 277 rice lines were planted in two field locations: Sanming City, Fujian Province and Sanya City, Hainan Province. The Sanming site was representative of a temperate climate and Sanya of a subtropical climate. The 22 -d-old seedlings were planted at a spacing of 17 × 25 cm in all field experiments. Nitrogen (N) fertilizer was applied at a level of 150 kg ha-1. Weeds and pests were controlled as locally recommended. Plant height (PH), tiller number (TN), panicle length (PL), thousand-grain weight (TGW), grain length (GL), grain width (GW), and the grain length-to-width ratio (L/W) were recorded at maturity. Three central plants in each plot were used to measure PH and TN. Then, the plants were harvested to measure grain traits. For TGW, GL, GW, and L/W, the data were collected using a SC-G rice grain appearance quality image analysis system (Wseen Detection Technology Co., Ltd, Hangzhou, China). Approximately 500 grains were spread on one 21 × 30 cm flat–bed surface, photographed using an Eloam high-speed photographic apparatus S500A3, and the image analysis system calculated GL, GW and L/W (Yin et al. 2015). Next, the grains were collected and a high precision balance (Amput, China) was used to weigh the grains and calculate TGW.
In addition, 200 g of grain from each line was harvested at Sanming in 2014 for grain quality analysis. AC, GT, GC and TPC were measured with three independent replications at the State Key Laboratory of Rice Biology, China National Rice Research Institute. GT and GC were measured according to the method of Tan (Tan et al. 1999).
For AC analysis, 50 mg of rice flour was placed in a 100 mL volumetric flask and 1 mL of ethanol (95 %) was added. The volumetric flask was shaken to disperse the rice flour and 9 mL of sodium hydroxide (1 mol/L) was added. The volumetric flask was placed in a boiling water bath for 10 min, and distilled water was added to make up 100 mL after cooling. The sample or standard sample, acetic acid (1 mol/L) and iodine/potassium iodide solution were mixed using an AA3 Continuous Flow Analyzer (SEAL Analytical GmbH, Germany). Absorbance data were collected and AC content was calculated using a standard curve. For TPC analysis, 200 mg of rice flour was placed in a digestion tube, 5 mL of concentrated sulfuric acid was added and the tube was placed in a digestion furnace (SEAL Analytical) for 1.5 h at 220 °C. Then, 2 mL of hydrogen peroxide was added and the tube was re-heated until the solution became colorless. The solution was transferred to a 100 mL volumetric flask and distilled water was added to make up 100 mL. The sample or standard, natrium salicylicum and sodium hypochlorite were mixed using an AA3 Continuous Flow Analyzer. Sodium nitroprusside was added to catalyze the reaction, generating a blue compound and 660 nm absorbance data were collected. The N content was calculated using a standard curve and TPC determined using the following formula:
where c is N content, v is volume, and w is sample weight.
Genotyping
A total of 193 SSRs distributed on the 12 chromosomes and another 34 InDel indica–japonica identification markers (Lu et al. 2009; Shen et al. 2004) were used in this study. Exactly 22, 27, 22, 11, 16, 15, 18, 22, 14, 10, 8, and 8 SSRs were mapped to chromosomes 1–2, respectively; and the corresponding average distances between SSR markers were 7.8, 7.6, 10.3, 14.8, 9.3, 9.2, 6.8, 6.1, 8.5, 11.3, 16.1, and 13.9 cM.
DNA was extracted using the CTAB method (Murray and Thompson 1980). The SSR and InDel markers were all amplified and separated by the following method. The PCR volume was 10 μL and the reaction was conducted on a 9700 PCR system (ABI) programmed as follows: 94 °C for 5 min; 30 cycles of 94 °C for 40 s, 55 °C for 40 s and 72 °C for 40 s; and 7 min of final extension at 72 °C. The amplified products were size-separated using 8 % polyacrylamide gel electrophoresis and detected by SYBR Green Gel Stain (Zeng et al. 2013). The alleles were recorded by the Imagine System (GeneSys, USA).
Data analysis
Phenotype profiling
The phenotype data were managed using Excel 2007 (MS Excel), and the descriptive statistics and variance analysis were produced using SPSS 17.0 (IBM SPSS). Broad-sense heritability was calculated using the formula: h 2 = σ 2 g /(σ 2 g + σ 2 e ), where \(\sigma^{2}_{g}\) is the genotypic variance and \(\sigma^{2}_{e}\) is the environmental variance (Wang et al. 2007).
Differentiation of indica–japonica rice
The indica gene frequency (Fi) and japonica gene frequency (Fj) were calculated using the method described by Lu et al. (Lu et al. 2009).
Genetic diversity
PowerMarker 3.25 (Liu and Muse 2005) was used to calculate the average number of alleles per locus, gene diversity (He), and the polymorphism information content (PIC) values.
Population structure
A subset of 90 SSR markers was chosen for population structure analysis, with a genetic distance far more than 10 cM between any two markers. The model-based program STRUCTURE 2.2 (Earl and vonHoldt 2012) was used to infer the population structure of the 277 rice accessions using a burn-in period of 10,000 and Monte Carlo Markov Chain replicates of 100,000. The number of groups (K) was set from 1 to 15, with seven independent runs each. Evanno’s method (Evanno et al. 2005) was used to calculate the most probable structure number for K. The rice lines were divided into different subgroups with a membership probability threshold (Q) of 0.80 (Zhang et al. 2011b). The admixed group (AD) consisted of those lines with Q < 0.80. PowerMarker was also used to calculate Nei’s genetic distance and to construct the Neighbor-Joining tree (Liu and Muse 2005). MEGA 4 (Tamura et al. 2007) was used to visualize the phylogenetic distribution tree for lines and groups. The main variation patterns for the alleles were generalized by principal coordinate analysis (PCoA) performed by NTSYSpc 2.10 (Rohlf 2002).
Linkage disequilibrium (LD) analysis
The software TASSEL 2.1 (http://www.maizegenetics.net/) was used to calculate the level of LD between pairs of loci both in the entire population and the indica population. For each SSR, the rare alleles with minor allele frequency less than 0.05 were pooled into one allelic class (Jin et al. 2010; Maccaferri et al. 2005). The relationship between LD decay and physical distance was estimated by a plot drawn using squared correlations of allele frequencies (r 2) against physical distance between linked loci.
Association analysis
The association analysis was undertaken using the mixed linear model (MLM) procedure in TASSEL 2.1 (http://www.maizegenetics.net/) (Bradbury et al. 2007). Since the phenotypic data were collected at two geographical locations in 2 years, we used best linear unbiased predictors (BLUPs) to integrate the data. The kinship was calculated using the software SPAGeDi (Hardy and Vekemans 2002). The model MLM_Q + K, using kinship and Q as the concomitant variable, was used to test association between genotypes and phenotypes. False discovery rate (FDR) was applied to identify statistically significant loci.
Identification of superior loci and alleles
Allele effects were estimated using the null allele method described by Breseghello (Breseghello and Sorrells 2006).
Results
Morphological analysis
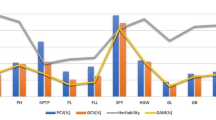
Seven yield traits were investigated at two geographical locations in 2013 and 2014 (Fig. 1). Statistical analysis demonstrated a high level of variation for all seven yield traits, with variable coefficients in the range of 9.81–30.02 %. The broad-sense heritability of the traits was >90 % except for TN, which was only 57.39 %, suggesting that TN was strongly affected by environment. Genotype had a highly significant effect (P < 0.01) for all seven traits (Table S2). Location had a significant effect for PH (P < 0.01), PL (P < 0.01) and L/W (P < 0.05). Year had a significant effect for PL (P < 0.05). Genotype × location had a significant effect for PH, PL and TGW (all P < 0.01). Genotype × year had no significant effects.
One nutritional and three cooking and eating quality characteristics were investigated at Sanming location in 2014 (Fig. 1). There was a wide variation in the four quality traits, with variable coefficients in the range of 12.19–35.53 %. Genotype had a highly significant effect for the four grain traits (P < 0.01) (Table S3).
Differentiation of indica–japonica rice
Among the 277 hybrid rice primal parental lines, 256 lines were classified as indica rice (92.42 %), 20 were japonica (7.22 %) and one was intermediate. The indica lines were further classified into 192 typical indica, 62 indica and two indica-clined based on the method (Lu et al. 2009). Similarly, the 20 japonica were classified into six japonica-clined, 10 japonica, and four typical japonica (Fig. S1 and Table S4).
Genetic diversity
By using the 193 SSR markers, 835 alleles were detected in the 277 rice lines in total. There were 2–15 alleles per locus, with an average of 4.326. The He was in the range of 0.014–0.881 with an average of 0.415. PIC was 0.014–0.869 with an average of 0.373. There were 792 and 478 alleles for indica and japonica, respectively (Table 1, Table S5).
Population structure
The model-based STRUCTURE 2.2 program was used to infer population structure. The 277 lines could be divided into two subgroups (Fig. 2a and b). With Q = 0.80, 253 indica lines were assigned to the subgroup POP 1, 20 japonica lines were assigned to subgroup POP 2 and four lines were assigned to AD (Fig. 2c). The genetic structure pattern was similar to the InDel marker classification (Table S4) where POP 1 included 192 typical indica and 61 indica lines. POP 2 included six japonica-clined, 10 japonica, and four typical japonica lines. The AD subgroup included one indica, two indica-clined and one intermediate line.
A neighbor-joining tree based on Nei’s distance was constructed (Fig. 3). The 277 rice lines were assigned to cluster I and II, which corresponded to POP 1 and POP 2, respectively. Clusters I and II contained indica and japonica rice, respectively. There were four sub-clusters in the indica cluster (POP 1A-D), which were related to landrace, maintainer, restorer, and lines from IRRI, respectively (Figs. 2d, 3).
PCoA based on the marker genotypes revealed two distinct clusters for the 277 lines (Fig. 4). The first and second principal components explained 13.22 and 7.11 % of the molecular variance, respectively.
These results indicated that the whole population included indica and japonica subgroups and that the indica subgroup was split into four different sub-subgroups.
LD and decay
The levels of LD between pairs of loci were inferred both in the entire population and the indica population. In the entire population, 64.57 % of the loci pairs were significant (P < 0.01) for LD, and 6.14 % of the loci pairs were both significant and syntenic. In the indica population, only 19.67 % of the loci pairs were significant (P < 0.01) for LD, but 7.91 % of the loci pairs were both significant and syntenic (Table S6). The minimum distance for LD decay was deduced via regression equation. LD decay distance of the entire and the indica population were ~2415 and ~118 kb, respectively (Fig. 5a, b), where the average pairwise correlation coefficient (r 2) dropped to 0.1.
Marker-trait association
Association analysis between SSR markers and 11 agronomic traits, including seven yield and four grain quality traits was carried out using MLM model over two years across two locations in the entire population and the 256 indica rice lines. However, 516 markers were significantly associated with 11 agronomic traits at the FDR correction level, or 353 were significant at the Benferroni correction (P < 0.05/n) level, in the entire population. The reasons for the many significant association markers were mainly a strong population structure and that LD decayed slowly in the entire population. Hence, we did not do more analysis across the entire population.
In the indica rice group, a total of 24 SSR markers were identified as significantly associated with six agronomic traits at the FDR correction level, explaining 1.03–7.21 % of the phenotypic variance. No significantly associated marker was detected for PH, PL, AC, GT and TPC at the FDR correction level. Therefore, we discarded all but the most significant markers for the five agronomic traits. In total, we identified 29 SSR markers associated with 11 agronomic traits (Table 2). Among the associated markers, 16 were located close to loci that had previously been identified.
RGS1 and RM6283 were co-associated with three grain shape traits, and they were located close to the known -loci GS3 and GL3.2/qGW-3. Moreover, four other markers (RM7187, RM6645, RM351 and RM1345) were co-associated with two grain shape traits. RM7187 was located close to d11, and RM351 close to GL7/GW7, while no known -locus was close to RM6645 and RM1345. More study on these genomic areas will be useful, as this could further our understanding of grain shape traits.
Identification of superior alleles and their carrier lines
The phenotypic effects of significant association marker alleles on agronomic traits and their carrier lines were analyzed (Table 3). For grain shape traits, including GL, GW, L/W and TGW, the alleles with the most positive effect were RM6283-93 (+0.19 mm), RM6283-96 (+0.40 mm), RM1345-128 (+0.63) and RM1370-173 (+0.67 g), from the carrier lines Minghui63, II-32B, Jinhangsimiao and Minghui86, respectively. The alleles RGS1-193 and RM6283-93 increased GL (+0.18 and +0.19 mm, respectively) and L/W (+0.13 and +0.11, respectively) but simultaneously reduced GW (−0.05 and −0.04 mm, respectively). The allele with the greatest negative effect for GW was RM1345-128 (−0.56 mm) from line Jinhangsimiao. The allele with the greatest negative effect for TN was RM3425-136 (−1.99) from Minghui86. For grain quality traits, the allele with the most positive effect for GC was RM331-167 (+5.59 mm) from R659. The allele RM411-110 reduced AC (−0.57 %), RM1022-148 increased GT (0.97) and RM495-165 increased TPC (0.06 %). The superior alleles identified here could be applied in marker assisted breeding practice to pyramid multiple favorable alleles into one plant.
Discussion
The differentiation of indica–japonica rice and the population structure of rice lines are important for parent matching in rice breeding. Population structure analysis is also necessary for association analysis. In previous studies, various rice population have been divided into different groups, for example the 203 USDA rice mini-core (URMC) subset was divided into five groups, AUS, IND, TEJ, TRJ, and ARO (Li et al. 2012); 905 worldwide rice cultivars were divided into five groups, IND, AUS, TEJ, TRJ, and INT (Huang et al. 2011); and 150 rice core local germplasm lines were divided into indica and japonica, with four subgroups in the indica group (Zhang et al. 2011b). In the present study, we used InDels and SSRs markers to assess indica–japonica properties and population structure in 277 hybrid primal parental lines. Our results assigned the entire population into japonica and indica groups, with indica further divided into four subgroups, representing landrace, maintainer, restorer and lines from IRRI. Our study and previous study (Zhang et al. 2011b) all indicated that the indica–japonica differentiation caused the observed population structure, and geographical distribution further promoted subgroup formation. The indica–japonica hybrid sterility is a common problem that prevents the use of heterosis between indica and japonica. According to our study, rice breeders could chose parental lines with appropriate gene frequencies Fi or Fj to avoid this disadvantage. Moreover, they could improve the restorer or maintainer lines by crossing them with landraces or rice lines from IRRI, or even with japonica, to extend heterosis and genetic diversity in future rice breeding programs. The clustering of heterotic groups has been successfully applied to maize breeding (Duvick 2005), so the elucidation of rice population structure will be of great importance to rice parent matching and to increase rice genetic diversity.
In the entire population, 64.57 % of the loci pairs were significant, but only 6.14 % loci pairs were both significant and syntenic. However, the percentages in the indica group were 19.67 and 7.91 %, respectively. Moreover, r 2 of the entire population was much higher than that of the indica population, especially for the loci pairs both significant and syntenic (Table S6). All of this indicated a strong population structure in the entire population lead higher LD level and much longer LD decay distance (Fig. 5a, b). Our results were similar with previous studies, 63 % of the SSR pairs in the 416 accessions were in LD, since the number of locus pairs in LD was reduced sharply within each subpopulation, with the SSR pairs in LD ranging from 5.9 to 22.9 % (Jin et al. 2010). Another study (Zhang et al. 2011b) found the LD decay distance were in the region of 40–50 cM in the entire germplasm set, <5 cM in the subgroups and ≦3 cM in the sub-subgroups.
Since a strong population structure in the entire population lead higher LD level and much longer LD decay distance, and most of the loci pairs (58.43 %) significant in LD but not on the same chromosome, there were so many markers significantly associated with 11 agronomic traits in the entire population, we regarded lots of the markers were as false positive. Moreover, the indica group represented 92.41 % of the entire population, and so we conducted association only in indica population.
Rice yield is a complex trait, determined by the number of panicles, number of grains per panicle, and grain weight (Xing and Zhang 2010). The number of panicles depends on the TN; the number of grains per panicle is related to PL, panicle branch number and setting percentage; and grain weight depends on GL, GW, and TGW. Grain milling quality, appearance quality, cooking and eating quality, and nutritional quality comprise overall grain quality, which is also a complex trait. In this study, the MLM model was used to analyze seven yield traits and four grain quality traits over 2 years at two locations. In total, 29 SSR markers were identified as associate with 11 traits. Among them, 16 were near known QTLs or genes. RGS1 and RM6283 were simultaneously associated with GL, GW and L/W. RGS1 was designed based on the GS3 sequence (Wang et al. 2011) and RM6283 is linked with GL3.2. GS3 is the major-effect QTL controller of grain weight and GL, but it is the minor-effect QTL controller of GW and grain plumpness (Mao et al. 2010). GL3.2 is a homolog of CYP78A13 and is associated with GL, GW, grain thickness, and TGW (Xu et al. 2014). RMw-513, RM3777 and RM1089 were associated with GW and were linked to GW5, which controls GW and grain weight (Yan et al. 2011). RM351 was associated with L/W and was linked to GL7/GW7, which was the main QTL for GL and GW. RM7187 was associated with GL and L/W and was linked to d11, which controlled PH; and over-expression of D11 increased GL, GW and TGW (Zhu et al. 2015). RM3700 was associated with L/W and was close to SG1. Over-expression of SG1 resulted in short grain and a dwarf phenotype (Zhu et al. 2015). RM6144 was associated with TN and was linked to OsCesA7. OsCesA7 participates in cellulose biosynthesis, and the cesa7 mutant exhibited brittle culms and other pleiotropic phenotypes including reduced TN (Wang et al. 2016). RM1307 was associated with TGW and was linked to TGW6, which encodes IAA-glucose hydrolase, affecting grain weight and enhancing rice yield (Ishimaru et al. 2013). The other 13 SSR markers have not been reported to be linked with any agronomic trait, and could be new loci for important agronomic trait.
Rice yield and grain quality are complex traits, and yield is significantly correlated with the number of superior alleles (Huang et al. 2015). Our study utilized the hybrid rice primal parental lines and intermediate materials collected from China and abroad, which included many important parental lines and intermediate materials for China’s hybrid rice such as Minghui 63, Minghui 86, Duoxi 1, Shuhui527 and II -32B. Our research grouped the primal parental lines of hybrid rice into different subgroups, identified significant loci associated with yield and grain quality traits, and analyzed the allele effect on phenotype. This information can be applied to parent selection for restorer or maintainer line improvement in three-line hybrid rice breeding. Furthermore, it will aid in marker assisted selection and molecular design breeding.
References
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Nishimura A, Angeles E, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309:741–745
Bradbury P, Zhang Z, Kroon D, Casstevens T, Ramdoss Y, Buckler E (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, Zhang W, Zhang L, Yu S, Wang G, Lian X, Luo J (2014) Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet 46:714–721
Duvick ND (2005) The contribution of breeding to yield advances in maize (Zea mays L.). Adv Agron 86:83–145
Earl D, vonHoldt B (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park J, Jane J, Miyao A, Hirochika H, Nakamura Y (2007) Characterization of SSIIIa-Deficient Mutants of Rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144:2009–2023
Gao Z, Zeng D, Cui X, Zhou Y, Yan M, Huang D, Li J, Qian Q (2003) Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci China Ser C 46:661–668
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, Li W, Guo Y, Deng L, Zhu C, Fan D, Lu Y, Weng Q, Liu K, Zhou T, Jing Y, Si L, Dong G, Huang T, Lu T, Feng Q, Qian Q, Li J, Han B (2011) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44:32–39
Huang X, Yang S, Gong J, Zhao Y, Feng Q, Gong H, Li W, Zhan Q, Cheng B, Xia J, Chen N, Hao Z, Liu K, Zhu C, Huang T, Zhao Q, Zhang L, Fan D, Zhou C, Lu Y, Weng Q, Wang Z-X, Li J, Han B (2015) Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun. doi:10.1038/ncomms7258
Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, Miyagawa H, Katoh E (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45:707–711
Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao J (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet 121:475–487
Li X, Yan W, Agrama H, Jia L, Jackson A, Moldenhauer K, Yeater K, McClung A, Wu D (2012) Unraveling the complex trait of harvest index with association mapping in rice (Oryza sativa L.). PLoS One. doi:10.1371/journal.pone.0029350
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Lu B, Cai X, Jin X (2009) Efficient indica and japonica rice identification based on the InDel molecular method: its implication in rice breeding and evolutionary research. Prog Nat Sci 19:1241–1252
Maccaferri Marco, Corinna Maria, Sanguineti Enrico Noli, Tuberosa R (2005) Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Mol Breed 15:271–289
Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA 107:19579–19584
Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Rohlf FJ (2002) NTSYS-PC: Numerical taxonomy and multivariate analysis system, version 2.10. Exeter Software, Setauket
Sato H, Suzuki Y, Sakai M, Imbe T (2002) Molecular characterization of Wx-mq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.). Breed Sci 52:131–135
Shen Y, Jiang H, Jin J, Zhang Z-B, Xi B, He Y, Wang G, Wang C, Qian L, Li X, Yu Q, Liu H, Chen D, Gao J, Huang H, Shi T, Yang Z (2004) Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol 135:1198–1205
Song X, Huang W, Shi M, Zhu M, Lin H (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Su Y, Rao Y, Hu S, Yang Y, Gao Z, Zhang G, Liu J, Hu J, Yan M, Dong G, Zhu L, Guo L, Qian Q, Zeng D (2011) Map-based cloning proves qGC-6, a major QTL for gel consistency of japonica/indica cross, responds by Waxy in rice (Oryza sativa L.). Theor Appl Genet 123:859–867
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan F, Li X, Yu B, Xing Z, Xu G, Zhang Q (1999) The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor Appl Genet 99:642–648
Wang L, Liu W, Xu Y, He Y, Luo L, Xing Y, Xu C, Zhang Q (2007) Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor Appl Genet 115:463–476
Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, Ma H, Zhang G, He Z (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40:1370–1374
Wang C, Chen S, Yu S (2011) Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice. Theor Appl Genet 122:905–913
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, Ye W, Meng X, Liu R, Chen H, Jing Y, Wang Y, Zhu X, Li J, Qian Q (2015) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47:944–948
Wang D, Qin Y, Fang J, Yuan S, Peng L, Zhao J, Li X (2016) A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PLoS One. doi:10.1371/journal.pone.0153993
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153:1747–1758
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Xu F, Fang J, Ou S, Gao S, Zhang F, Du L, Xiao Y, Wang H, Sun X, Chu J, Wang G, Chu C (2014) Variations in CYP78A13 coding region influence grain size and yield in rice. Plant, Cell Environ 38:800–811
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yan S, Zou G, Li S, Wang H, Liu H, Zhai G, Guo P, Song H, Yan C, Tao Y (2011) Seed size is determined by the combinations of the genes controlling different seed characteristics in rice. Theor Appl Genet 123:1173–1181
Yin C, Li H, Li S, Xu L, Zhao Z, Wang J (2015) Genetic dissection on rice grain shape by the two-dimensional image analysis in one japonica × indica population consisting of recombinant inbred lines. Theor Appl Genet 128:1969–1986
Zeng Y, Zhu Y, Lian L, Xie H, Zhang J, Xie H (2013) Genetic analysis and fine mapping of the pubescence gene GL6 in rice (Oryza sativa L.). Chin Sci Bull 58:2992–2999
Zhang G, Cheng Z, Zhang X, Guo X, Su N, Jiang L, Mao L, Wan J (2011a) Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 54:448–459
Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y (2011b) Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS One. doi:10.1371/journal.pone.0027565
Zhao K, Tung C, Eizenga G, Wright M, Ali ML, Price A, Norton G, Islam M, Reynolds A, Mezey J, McClung A, Bustamante C, McCouch S (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. doi:10.1038/ncomms1467
Zhu X, Liang W, Cui X, Chen M, Yin C, Luo Z, Zhu J, Lucas W, Wang Z, Zhang D (2015) Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J 82:570–581
Acknowledgments
We thank Prof. Chenwu Xu and Pengcheng Li at Agricultural College, Yangzhou University for technical support on statistics. This work was supported by grants from the National Program on the Development of Basic Research of China (Grant No. 2013CBA01405-7), the Hi-tech Research and Development (863) Program of China (Grant Nos. 2014AA10A603 and 2014AA10A604), the Special Foundation of Non-Profit Research Institutes of Fujian Province (Grant Nos. 2014R1021-15 and 2015R1021-8) and the Youth Science Fund of the Fujian Academy of Agriculture (Grant No. 2012DQA-4).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, Y., Zheng, Y., Cai, Q. et al. Population structure and association analysis of yield and grain quality traits in hybrid rice primal parental lines. Euphytica 212, 261–273 (2016). https://doi.org/10.1007/s10681-016-1766-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-016-1766-3