Abstract
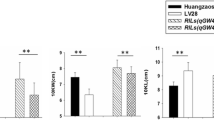
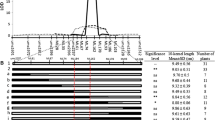
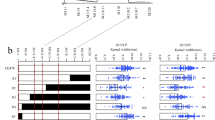
Kernel weight (KWEI), a complex quantitative trait that is largely determined by kernel size, is a key component of grain yield in maize. In our previous studies, qKW7, a putative pleiotropic QTL associated with both KWEI and kernel width (KWID), was detected within the 7.02 bin region on chromosome 7 by both F2:3 population and recombinant inbred line (RIL) population from a Ye478 and Huangzaosi cross. Therefore, a series of backcross populations from the same cross have been developed for fine mapping qKW7. By using a large BC3F4 segregation population, the qKW7 QTL was mapped to a physical interval of 647 Kb with several genes. An association panel with 627 diverse inbred lines was used to process the association analysis, and the results indicated that three KWEI- and KWID-related quantitative trait nucleotides (QTNs) were tightly linked to qKW7. Sequence analysis revealed that all three detected QTNs were located within the gene region of GRMZM2G114706, which encodes an ankyrin protein kinase that regulates endosperm development. Finally, the putative genes responsible for qKW7 are discussed.





Similar content being viewed by others
References
Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5:22–29
Alvarez PS, López CG, Gambín BL, Abertondo VJ, Borrás L (2013) Dissecting the genetic basis of physiological processes determining maize kernel weight using the IBM (B73 × Mo17) Syn4 population. Field Crop Res 145:33–43
Austin DF, Lee M (1996) Comparative mapping in F2:3 and F6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor Appl Genet 92:817–826
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Berger F (2003) Endosperm: the crossroad of seed development. Curr Opin Plant Biol 6:42–50
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Burland TG (1999) DNASTAR’s Lasergene sequence analysis software. Bioinform Methods Protoc 132:71–91
Chen JY, Guo L, Ma H, Chen YY, Zhang HW, Ying JZ, Zhuang JY (2014) Fine mapping of qHd1, a minor heading date QTL with pleiotropism for yield traits in rice (Oryza sativa L.). Theor Appl Genet 127:2515–2524
Cheng WH, Taliercio EW, Chourey PS (1996) The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endospermand maternal cells in the pedicel. Plant Cell 8:971–983
Fan CH, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Finnegan EJ, Peacock WJ, Dennis ES (2000) DNA methylation, a key regulator of plant development and other processes. Curr Opin Genet Dev 10:217–223
Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324:1447–1451
Gupta PK, Rustgi S, Kumar N (2006) Genetic and molecular basis of grain size and grain number and its relevance to grain productivity in higher plants. Genome 49:565–571
Huang XH, Wei XH, Sang T, Zhao QA, Feng Q, Zhao Y, Li CY, Zhu CR, Lu TT, Zhang ZW, Li M, Fan DL, Guo YL, Wang A, Wang L, Deng LW, Li WJ, Lu YQ, Weng QJ, Liu KY, Huang T, Zhou TY, Jing YF, Li W, Lin Z, Buckler ES, Qian QA, Zhang QF, Li JY, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Jiang PS, Zhang HX, Lv XL, Hao CF, Li B, Li MS, Wang HW, Ci XK, Zhang SH, Li XH, Shi ZS, Weng JF (2013) Analysis of meta-QTL and candidate genes related to yield components in maize. Acta Agron Sin 39:969–978
Kapazoglou A, Tondelli A, Papaefthimiou D, Ampatzidou H, Francia E, Stanca MA, Bladenopoulos K, Tsaftaris AS (2010) Epigenetic chromatin modifiers in barley: IV. The study of barley polycomb group (PcG) genes during seed development and in response to external ABA. BMC Plant Biol 10:73
Kesavan M, Song JT, Seo HS (2013) Seed size: a priority trait in cereal crops. Physiol Plantarum 147:113–120
Li HH, Ye GY, Wang JK (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Li Q, Li L, Yang XH, Warburton ML, Bai GH, Dai JR, Li JS, Yan JB (2010a) Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol 10:143
Li Q, Yang XH, Bai GH, Warburton ML, Mahuku G, Gore M, Dai JR, Li JS, Yan JB (2010b) Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor Appl Genet 120:753–763
Li Y, Ma XL, Wang TY, Li YX, Liu C, Liu ZZ, Sun BC, Shi YS, Song YC, Carlone M, Bubeck D, Bhardwaj H, Whitaker D, Wilson W, Jones E, Wright K, Sun SK, Niebur W, Smith S (2011) Increasing maize productivity in China by planting hybrids with germplasm that responds favorably to higher planting densities. Crop Sci 51:2391–2400
Li YL, Yang ML, Dong YB, Wang QL, Zhou YG, Zhou Q, Shen BT, Zhang FF, Liang XJ (2012) Three main genetic regions for grain development revealed through QTL detection and meta-analysis in maize. Mol Breed 30:195–211
Li CH, Li YX, Sun BC, Peng B, Liu C, Liu ZZ, Yang ZZ, Li QC, Tan WW, Zhang Y, Wang D, Shi YS, Song YC, Wang TY, Li Y (2013a) Quantitative trait loci mapping for yield components and kernel-related traits in multiple connected RIL populations in maize. Euphytica 193:303–316
Li H, Peng ZY, Yang XH, Wang WD, Fu JJ, Wang JH, Han YJ, Chai YC, Guo TT, Yang N, Liu J, Warburton ML, Cheng YB, Hao XM, Zhang P, Zhao JY, Liu YJ, Wang GY, Li JS, Yan JB (2013b) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45:43–50
Li YX, Li Y, Ma XL, Liu C, Liu ZZ, Tan XJ, Sun BC, Shi YS, Song YC, Wang TY, Smith S (2014) Contributions of parental inbreds and heterosis to morphology and yield of single-cross maize hybrids in China. Crop Sci 54:76–88
Liu RX, Jia HT, Cao XL, Huang J, Li F, Tao YS, Qiu FZ, Zheng YL, Zhang ZX (2012a) Fine mapping and candidate gene prediction of a pleiotropic quantitative trait locus for yield-related trait in Zea mays. PLoS ONE 7:e49836
Liu ZZ, Wu X, Liu HL, Li YX, Li QC, Wang FG, Shi YS, Song YC, Song WB, Zhao JR, Lai JS, Li Y, Wang TY (2012b) Genetic diversity and population structure of important Chinese maize inbred lines revealed by 40 core simple sequence repeats (SSRs). Sci Agric Sin 45:2107–2138
Liu Y, Wang LW, Sun CL, Zhang ZX, Zheng YL, Qiu FZ (2014) Genetic analysis and major QTL detection for maize kernel size and weight in multi-environments. Theor Appl Genet 127:1019–1037
Lu YL, Yan JB, Guimaraes CT, Taba S, Hao ZF, Gao SB, Chen SJ, Li JS, Zhang SH, Vivek BS, Magorokosho C, Mugo S, Makumbi D, Parentoni SN, Shah T, Rong TZ, Crouch JH, Xu YB (2009) Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms. Theor Appl Genet 120:93–115
Maitz M, Santandrea G, Zhang ZY, Lal S, Hannah LC, Salamini F, Thompson RD (2000) rgf1, a mutation reducing grain filling in maize through effects on basal endosperm and pedicel development. Plant J 23:29–42
Mao HL, Sun SY, Yao JL, Wang CR, Yu SB, Xu CG, Li XH, Zhang QF (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA 107:19579–19584
Marmagne A, Rouet MA, Ferro M, Rolland N, Alcon C, Joyard J, Garin J, Barbier-Brygoo H, Ephritikhine G (2004) Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteom 3:675–691
Monaco MK, Stein J, Naithani S, Wei S, Dharmawardhana P, Kumari S, Amarasinghe V, Youens-Clark K, Thomason J, Preece J, Pasternak S, Olson A, Jiao YP, Lu ZY, Bolser D, Kerhornou A, Staines D, Walts B, Wu GM, D’Eustachio P, Haw R, Croft D, Kersey PJ, Stein L, Jaiswal P, Ware D (2014) Gramene 2013: comparative plant genomics resources. Nucl Acids Res 42:D1193–D1199
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326
Patrick JW, Offler CE (2001) Compartmentation of transport and transfer events in developing seeds. J Exp Bot 52:551–564
Peng B, Wang Y, Li YX, Liu C, Zhang Y, Liu ZZ, Tan WW, Wang D, Sun BC, Shi YS, Song YC, Wang TY, Li Y (2010) Correlation analysis and conditional QTL analysis of grain yield and yield components in maize. Acta Agron Sin 36:1624–1633
Peng B, Li YX, Wang Y, Liu C, Liu ZZ, Tan WW, Zhang Y, Wang D, Shi YS, Sun BC, Song YC, Wang TY, Li Y (2011) QTL analysis for yield components and kernel-related traits in maize across multi-environments. Theor Appl Genet 122:1305–1320
Peng B, Li YX, Wang Y, Liu C, Liu ZZ, Zhang Y, Tan WW, Wang D, Shi YS, Sun BC, Song YC, Wang TY, Li Y (2013) Correlations and comparisons of quantitative trait loci with family per se and testcross performance for grain yield and related traits in maize. Theor Appl Genet 126:773–789
Prioul JL, Jeannette E, Reyss A, Gregory N, Giroux M, Hannah LC, Causse M (1994) Expression of ADP-glucose pyrophosphorylase in maize (Zea mays L.) grain and source leaf during grain filling. Plant Physiol 104:179–187
Salvi S, Tuberosa R (2005) To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci 10:297–304
SanMiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplication of intergene retrotransposons. Ann Bot 82:37–44
Sekhon RS, Lin HN, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66:553–563
Sheridan WF, Neuffer MG (1980) Defective kernel mutants of maize II. Morphological and embryo culture studies. Genetics 95:945–960
Shi JQ, Huang SM, Zhan JP, Yu JY, Wang XF, Hua W, Liu SY, Liu GH, Wang HZ (2014) Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res 21:53–68
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40:1023–1028
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Sosso D, Mbelo S, Vernoud V, Gendrot G, Dedieu A, Chambrier P, Dauzat M, Heurtevin L, Guyon V, Takenaka M, Rogowsky PM (2012) PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing mitochondrion. Plant Cell 24:676–691
Su ZQ, Hao CY, Wang LF, Dong YC, Zhang XY (2011) Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor Appl Genet 122:211–223
Wang M, Yan JB, Zhao JR, Song W, Zhang XB, Xiao YN, Zheng YL (2012a) Genome-wide association study (GWAS) of resistance to head smut in maize. Plant Sci 196:125–131
Wang SK, Wu K, Yuan QB, Liu XY, Liu ZB, Lin XY, Zeng RZ, Zhu HT, Dong GJ, Qian Q, Zhang GQ, Fu XD (2012b) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Wang YJ, Huang ZJ, Deng DX, Ding HD, Zhang R, Wang SX, Bian YL, Yin ZT, Xu XM (2013) Meta-analysis combined with syntenic metaQTL mining dissects candidate loci for maize yield. Mol Breed 31:601–614
Weig A, Franz J, Sauer N (1994) Isolation of a family of cDNA clones from Ricinus communis L. with close homology to the hexose carriers. J Plant Physiol 143:178–183
Weng JF, Gu SH, Wan XY, Gao H, Guo T, Su N, Lei CL, Zhang X, Cheng ZJ, Guo XP, Wang JL, Jiang L, Zhai HQ, Wan JM (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res 18:1199–1209
Williams LE, Lemoine R, Sauer N (2000) Sugar transporters in higher plants—a diversity of roles and complex regulation. Trends Plant Sci 5:283–290
Wu X, Li YX, Shi YS, Song YC, Wang TY, Huang YB, Li Y (2014) Fine genetic characterization of elite maize germplasm using high-throughput SNP genotyping. Theor Appl Genet 127:621–631
Yan JB, Shah T, Warburton ML, Buckler ES, McMullen MD, Crouch J (2009) Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 4:e8451
Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, Zhang QF (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 4:319–330
Zhang ZH, Liu ZH, Hu YM, Li WH, Fu ZY, Ding D, Li HC, Qiao MM, Tang JH (2014) QTL analysis of kernel-related traits in maize using an immortalized F2 population. PLoS ONE 9:e89645
Acknowledgments
This work was partially supported by grants provided by the National Natural Science Foundation of China (91335206), the Programmes of MST and MOA of China (2011CB100105, 2014CB138200, 201303007), and the CAAS (Innovation Program). We greatly thank Professor Jinsheng Lai of the China Agricultural University for sharing re-sequence information for marker development in this study.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
10681_2016_1706_MOESM1_ESM.tif
Linkage analysis of the major QTL for KWEI and KWID on chromosome 7 in the RIL population of Ye478×HZS. The numbers adjacent markers represent their genetic position (cM). Supplementary material 1 (TIFF 88 kb)
10681_2016_1706_MOESM2_ESM.tif
The comparisons of phenotypic variations of qKW7 alleles between HZS genotype individuals and Ye478 genotype individuals for KWEI (a) and KWID (b) in RIL population. The P-values of the t-test are noted in the right corner below. Supplementary material 2 (TIFF 62 kb)
10681_2016_1706_MOESM3_ESM.tif
Summary statistics of kernel related traits. a), correlation analysis of KWEI among environments across two years. Supplementary material 3 (TIFF 289 kb)
10681_2016_1706_MOESM4_ESM.tif
Summary statistics of kernel related traits. b), correlation analysis of KWID among environments across two years. Supplementary material 4 (TIFF 267 kb)
10681_2016_1706_MOESM5_ESM.tif
Summary statistics of kernel related traits. c), correlation analysis of KWEI and KWID under five environments. In order to picture the simple correlation between environments, missing entries were imputed with average values across all environments. KWEI means the 100-kernel weight. KWID means 10-kernel width. ‘11’ means 2011 year, ‘12’ means 2012 year. BJ is an abbreviation of Beijing, CC is an abbreviation of Changchun in Jilin province, NC is an abbreviation of Nanchong in Sichuan province, TA is an abbreviation of Tai’an in Shandong province, XX is an abbreviation of Xinxiang in Henan province. Histograms show the distribution of KWEI and KWID. Numbers in the up-right triangle are the Pearson Coefficient among different environment pairs. The low-left triangle shows the scatter plots among environment pairs. Supplementary material 5 (TIFF 290 kb)
Rights and permissions
About this article
Cite this article
Li, X., Li, Yx., Chen, L. et al. Fine mapping of qKW7, a major QTL for kernel weight and kernel width in maize, confirmed by the combined analytic approaches of linkage and association analysis. Euphytica 210, 221–232 (2016). https://doi.org/10.1007/s10681-016-1706-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-016-1706-2