Abstract
To understand the molecular basis of potato starch related traits and the underlying starch biosynthesis and degradation, a Quantitative Trait Locus (QTL) analysis in combination with a candidate gene approach was performed. The diploid mapping population C × E, consisting of 249 individuals, was assayed over two consecutive years, for chipping colour, cold induced sweetening, starch content, starch granule size, starch gelling temperature, starch enthalpy, amylose content and degree of starch phosphorylation. QTLs were observed for all traits, except enthalpy on eight out of the twelve potato chromosomes. Several QTLs were found to be consistent over 2 years. Clustering of co-localizing QTLs was observed on some chromosomes, indicating common genetic factors for the different traits. On chromosome 2, Soluble Starch Synthase 2 mapped on the same position as QTLs for starch phosphorylation, starch gelling temperature and amylose content. α-glucan, water dikinase co-localizes on chromosome 5 together with QTLs for starch phosphorylation and cold induced sweetening. Furthermore, the genes coding for two phosphorylases (StPho1a and StPho2) coincide with QTLs for starch gelling temperature, chipping colour and starch granule size on chromosome 2 and a QTL for starch phosphorylation on chromosome 9, respectively. The results suggest allelic variation acting on the genetics of the different traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Starch is the main insoluble carbohydrate in plants and is present in many different plant organs, as transitory starch in green leaves or as storage starch in roots, tubers and seeds (Kossmann and Lloyd 2000). In potato, starch is mainly accumulated in starch granules of tubers and consists of two different polysaccharides, amylose and amylopectin in a 1:4 ratio respectively. This ratio has already been found to correlate with a number of different starch characteristics such as retro gradation of starch pastes (Visser et al. 1997a), poor transparency of starch gels and low adhesiveness (Visser et al. 1997b) and high granule melting temperatures (Schwall et al. 2000). Other starch properties including the degree of branching of amylopectin (Kortstee et al. 1998) and the phosphate contents of starch, can also have a profound influence on the characteristics and practical application of starch. Starch properties do not only have an impact on physio–chemical characteristics of starch as gelling agent, they also influence frying quality due to cold induced sweetening of potato tubers during cold storage in a direct manner. During cold induced sweetening, starch is broken down to the sugars glucose and fructose, making the tubers unsuitable for processing (Burton 1969). The degree of starch phosphorylation has been reported to influence the amount of cold induced sweetening (Lorberth et al. 1998).
In the gene pool of potato a wide natural variation is present with respect to different features of starch, although certain properties can only be achieved through chemical modification. Significant effort has already been put into elucidating the complete biosynthetic pathway of starch granules and understanding the mechanisms underlying the observed variation. Most of the enzymes involved in assembling starch granules are well known. A simplified scheme of starch metabolism is shown in Fig. 1. The substrate for the biosynthesis of starch polymers, ADP-glucose, is produced from glucose-6-P via glucose-1-P by phosphoglucomutase and ADP glucose pyrophosphorylase (AGPase) respectively (Mullerrober et al. 1992; Stark et al. 1992). ADP glucose is then used by the different starch synthase isoforms to construct the starch polymers. Starch synthases I, II and III (SSSI, SSSII and SSSIII) are mainly present in the soluble phase whereas the granule bound starch synthase I (GBSSI) is bound to the granule and catalyses the synthesis of amylose (Kuipers et al. 1994). Other enzymes such as branching enzymes (SBE), debranching enzymes (DBE) and phosphorylating enzymes as α-glucan, water dikinase (GWD) complete the synthesis of starch granules (Hofvander et al. 2004; Ritte et al. 2002). The first step is the break down of the starch back to the glucoses by AGPases, Starch Phosphorylases (StPho) and amylases, followed by the breakdown of glucose to sucrose by UDP glucose pyrophosphorylase (UGPase) and sucrose-6-phosphate synthase (SPS) and then the final step from sucrose to the reducing sugars glucose and fructose by invertases (Sowokinos 2001).
Simplified schematic overview of the synthesis and degradation of starch within and outside the amyloplast showing the following enzymes: UGPase, UDP-glucose pyrophosphorylase; PGM, Phosphoglucomutase; SUS, sucrose synthase; Inv, Invertase; StUCP, starch uncoupling protein; AGPase, ADP-glucose pyrophosphorylase; SSS, Soluble starch synthase; StPho, starch phosphorylase; GWD, α-glucan, water dikinase; GBSS, Granule bound starch synthase starch synthase
Different sets of genes control the synthesis and the degradation of starch granules and influence potato quality traits. To analyse the influence of these genes on phenotypic variation for starch properties, chipping colour and cold induced sweetening Quantitative Trait Locus (QTL) analysis is an appropriate method. Almost all genes involved have been mapped and cloned, creating the opportunity to strengthen QTL analysis by including candidate gene markers in the analysis. Here we describe the QTL analysis for a number of novel starch characteristics such as starch granule size, melting temperature and enthalpy, degree of phosphorylation, amylose content and overall starch content, as well as for chipping colour and the impact of cold induced sweetening on chipping colour. By including markers for the majority of starch related genes into the analysis we were not only able to identify the QTLs, but also indicate which genes are putatively involved in the different traits. Furthermore, by correlation analysis of the phenotypic data and comparison of the QTL mapping results, correlated characteristics can be identified.
Materials and methods
Plant material and linkage map construction
The diploid potato mapping population C × E was used, descending from the backcross between C (US-W5337.3) (Hannemann and Peloquin 1967) and E (77.2102.37) where C is the recurrent parent (Jacobsen 1980). In this study an expanded version of the linkage map of van Eck et al. (1995) was used, with an offspring size expanded to 249 full sib descendants. All additional individuals were genotyped using 16 AFLP markers with 15 primer combinations E32/M47, E32/M48, E32/M49, E32/M50, E32/M51, E32/M54, E32/M59, E32/M61, E35/M47, E35/M48, E35/M50, E38/M59, E39/M60, E45/M59 and E45/M60. The P-locus represents a morphological marker involved in blue anthocyanin pigmentation. Furthermore, the SSR markers STM0003, STM0019, STM0037, STM0051, STM1100, STM2005, STM2028, STM3012, STM3016, STM3023, STM5127, STM5136, STM5148 (Reid et al. 2011), and StI001, StI003, StI007, StI022, StI024, StI028, StI029, StI032, StI033, StI046, StI051, StI053 and StI057 (Feingold et al. 2005) were used. SSR markers were amplified in separate PCR reactions in a 20 μl reaction volume, containing 10 ng genomic DNA, 75 mM Tris–HCl pH 9.0, 20 mM (NH4)2SO4 0.1 % (w/v) Tween 20, 2.5 mM MgCl2, 100 μM of each dNTP (Fermentas), 4 pmol of each primer and 0.3U Goldstar Taq DNA polymerase (Eurogentec). The PCR cycles were 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 45 s and a final extension at 72 °C for 10 min. Differently labelled PCR products (6-FAM, HEX and NED) were combined in appropriate amounts to obtain optimal peak patterns for detection. The fluorescently labelled products were separated by capillary electrophoresis using an ABI PRISM 3700 DNA Analyzer (Applied Biosystems). Electropherograms were created automatically using GeneScan Analysis Software v3.7 (Applied Biosystems). Peak mobilities and areas were determined using ABI PRISM Genotyper ® 3.6 NT software. Genes involved in carbohydrate metabolism were mapped as SCAR or CAPS markers are shown in Table 1.
The integrated map of the C × E population consisting of 192 dominantly scored AFLP markers, 28 SSR markers and 24 CAPS/SCAR markers calculated as done previously (van Eck et al. 1995). The linkage groups were assigned to the 12 linkage groups representing the 12 chromosomes on the basis of the earlier version of this map comprising a total length of 996 cm.
All clones were grown in field trials on clay soil near Wageningen, the Netherlands during the normal potato-growing season (April–September). Each genotype was planted in four-hill plots, in three replications with randomised plots. The field trials were repeated over 4 years. The data on potato chipping colour were obtained on the harvests of the years 1998 and 1999, the different starch characteristics were measured on the harvests of the years 2002 and 2003.
Analysis of potato tuber starch characteristics
Directly after harvest of a single plot, 0.5 kg of fresh potato tubers per genotype was peeled, sliced and mixed and grinded. During grinding of the tuber slices, the juice was collected and 0.01 % of sodium metabisulphite (Na2S2O5) was added. Collected juices were taken to 4 °C to settle for at least 3 h. The juice was separated from the settled starch and demineralised water was added. The starch was mixed with the water and again given the opportunity to settle at 4 °C for 3 h, this washing step was repeated two more times. Finally the starch was poured on a Whatman filter paper on a Buchner funnel and later air dried at room temperature before being powdered using a sieve shaker (Retsch, Germany).
All the starch characteristics were analysed in three technical repeats of the same starch sample from a single field plot. The starch samples from the harvests of the years 2002 and 2003 essentially represent biological replications, and the final trait value is an average over these three repeats and 2 years. The starch granule sizes of all samples were measured using a Coulter Counter Multisizer II (Beckman-Coulter, High Wycombe, UK) using a orifice tube. Approximately 10 mg of starch was suspended in about 150 ml of isotonic solution. The measurements of the granule sizes were each time recorded for at least 50,000 particles.
The parameters onset temperature of melting (TO) and differences in enthalpy (ΔH) were measured automatically by Differential Scanning Calorimetry (DSC), using the Perkin-Elmer Pyrus 6 (Perkin-Elmer, Vlaardingen, the Netherlands). The characteristics were automatically measured with the accompanying software. The experiment was done as described by (Ji et al. 2003).
Measuring the degree of starch phosphorylation (in nmol PO4/mg starch) was done using a high throughput 96-well format. 20 mg of starch was put in a glass tube, together with 250 μl 70 % HClO4 and heated at 250 °C for 25 min. Then 50 μl 30 % H2O2 was added and heated at 250 °C for another 5 min. After cooling down and the volume was increased to 2 ml by adding H2O. 100 μl of the starch sample was then put into a 96-well microtiter plate and 200 μl of colour reagent (0.75 % (NH4)6Mo7O24·4H2O, 3 % FeSO4·7H2O and 0.75 % SDS dissolved in 0.375 M H2SO4) was added and mixed for 9 s in the microtiter plate reader. After incubation for 10 min at room temperature, the absorbance was measured at 750 nm in the microtiter plate reader against a reference sample. The amount of nmol PO4 per mg starch could be calculated using the following formula: nmol PO4/mg starch = (C microtiter plate × 20)/(Weight × dw%/100)), with “C microtiter plate being the temperature in °C, “weight” the amount of starch in mg and “dw%” the percentage dry weight of the starch.
Amylose content was determined using the protocol by (Hovenkamp-Hermelink et al. 1988), by extracting the starch with perchloric acid followed by determination of the absorption at 618 and 550 nm after staining with Lugol’s solution (I2-KI).
Starch content (also referred to as underwater weight) was determined for each plot in 1998 and 1999 by calculating a measure for it based on the dry and under water weight. The formula used was (5000/dry weight (g)) × underwater weight (g).
Determination of potato chipping colour
The potatoes of each plot of the three replications were harvested and then divided into three equal batches, which makes nine batches per genotype. The three replications of the first batch of potatoes was fried in 2 weeks after harvest, two other batches were stored in crates in a climate room for 3 months at 4 °C. The three replicates of the second batch of potatoes was fried directly after the 3 months storage. The three replicates of the third batch was reconditioned at room temperature for 3 weeks before frying, to see if the potatoes could recover from the cold storage treatment.
For the frying of the potatoes, three slices of three potato tubers were taken and fried in peanut oil at 180 °C until the slices showed no bubbling anymore. Frying colour was visually assessed on a scale from 1 (dark) to 9 (light). The trait values for cold induced sweetening are based on the differences in frying colour observed immediately after harvest and cold storage. The trait values for the reconditioning are based on the differences in frying colour observed immediately after cold storage and after reconditioning.
QTL mapping
The phenotypic data were tested for normality and when not normally distributed, transformed (square root). QTL analysis was performed using the software package MapQTL® version 5.0 (Van Ooijen 2009). QTL analysis was done using the interval mapping method developed by (Lander and Botstein 1989). The results of the interval mapping were compared to the results of the Kruskal–Wallis test and no differences between the results of the two analyses were observed. Detection of a QTL was also done using a LOD threshold calculated with the permutation test option provided in MapQTL® (Van Ooijen 2009). The two-LOD support interval was taken as a confidence interval for a QTL.
To estimate the cultivar means Yijk for chipping colour across years the following model was used. Y ijk = μ + Genotype i + yearj + (replicate within year)k(j) + e ijk , where (replicate within year) implies the three replicated plots within a field trial from which phenotypes were recorded. The cultivar means for starch phenotypes were obtained one a per year basis using Y ijk = μ + genotype i + replicatej + e ij , where replicate within the year of the field trial implies the three technical replicates based on one starch sample. Random terms are underlined. Heritabilities for cultivar means across 2 years and three plot replicates were calculated according to: h 2 = σ 2g /(σ 2g + σ 2y /2 + σ 2e /3), with σ 2g the variance component for cultivar main effects, σ 2y the variance component for the year effect and σ 2e the error variance. The genotypic main effects from the above mixed model was used to inspect trait correlations.
Results
Phenotypic data analysis
The starch characteristics were analysed over 2 years and averaged per year over three technical repeats. Figure 2 shows the distributions per year for the different starch traits. All traits values were normally distributed with the exception of the 2002 data for enthalpy. The segregation of the trait values was transgressive in both years (Fig. 2). In the cases of starch phosphorylation, onset temperature of starch gelling, enthalpy, granule size and percentage amylose, the parental values of C and E are in the middle of the range of the values of the progeny. The level of observed variation of the traits was similar between years, however, we observed a shift in range of the data between years. For instance, the average amount of PO4 per gram starch was 4.77 mg higher in 2002 than in 2003 but the range of the observations for both years was 25 mg PO4 (the difference between the highest and lowest of the observed values in each year). A similar shift in range of data was also observed for most of the other starch traits. For the data of 2002 starch phosphorylation, granule size and percentage amylose was on average higher than in 2003, 4.77 mg PO4, with 1.3 μm and 1.3 % respectively. Onset temperature of starch gelling and enthalpy was higher in 2003, at 0.7 and 1.0 °C respectively. The level of variation in the data is similar in both years for starch phosphorylation being 25 mg PO4, and an average granule size of 12 μm. Very high correlations between years can be found for starch phosphorylation, starch gelling temperature and starch content, with correlation coefficients of 0.88, 0.79 and 0.72 respectively. Enthalpy, average granule size and percentage of amylose are much less stable between years, with correlation scores of −0.46, 0.47 and 0.45 respectively.
Distribution of the different starch characteristics in the C × E population. The white bars represent the data from 2002 and the black bars the data from 2003, or in the case of starch content 1998 and 1999 respectively. The letters C and E represent the parental values for the individual years. Parental values for C for 2002 are not present due to absence of starch samples
Not only correlations between years within a trait could be calculated but also the correlations between different traits were calculated and the significance of the correlation, shown by the Pearson correlation coefficient and the corresponding p values (Table 2). Several starch characters showed significant correlation coefficients (p < 0.001). The degree of starch phosphorylation is correlated with starch gelling temperature, average granule size and percentage amylose, with correlation coefficients of 0.42, −0.42 and −0.55. Also, granule size and percentage amylose are significantly correlated with a score of 0.34. Based on this data, we can conclude that several of the starch characteristics have an effect on one another.
Chipping colour and cold induced sweetening were scored over 2 years. The phenotypic data was similar in both years (ρ = 0.72, p < 0.0001). A preliminary QTL analysis showed near complete coincidence of QTLs (data not shown), which justified averaging across years. The variation within the C × E population with regard to chipping colour is very large. On the scale of 1–9, progeny genotypes exhibit scores ranging from 1 to 8. Photographs of the different degrees of discolouration are shown in Fig. 3a. The distributions of the chipping colour scores, at the three different time points, as well as those of the values for cold induced sweetening are given in Fig. 3b. Just as for the starch characteristics, the chipping colour has an approximately normal distribution and a transgressive segregation at all three time points, see Fig. 3b. Broad sense heritabilities are estimated to be 0.77, 0.71 and 0.68 for chipping colour directly after harvest, after 3 months storage at 4 °C and reconditioning at room temperature respectively. For cold induced sweetening, the heritabilities range from 0.69 for the difference between chipping colour after harvest and after storage, to 0.66 for the difference between the after harvest and after reconditioning time points. In order to establish a relationship between different chipping colours at the different time points and cold induced sweetening, correlation coefficients with associated p values were calculated and are presented in Table 2. Significant correlations (p < 0.001) are found between chipping colour directly after harvest and after reconditioning and between chipping colour after 4 °C and reconditioning. With regard to cold induced sweetening, several significant correlations are observed (Table 2). These correlations exist between chipping colour at different time points as well as between several differences between these time points that we use as an indication for cold induced sweetening.
a Photographs of the different chipping colours, ranging from 1 (dark) to 9 (light). b Distribution of the phenotypic values for chipping colour ranging from 1 (dark) to 9 (light), and c Distribution of the phenotypic values for cold induced sweetening. For chipping colour (b) the white bars represent the phenotypic values directly after harvest, the grey bars represent the phenotypic values after 4 °C storage and the black bars represent the phenotypic values after reconditioning at room temperature. In c the white bars represent the population distribution of the severity of cold induced sweetening, measured as the (on average five points) difference between trait values measured directly after harvest and storage at 4 °C. The grey bars represent the population distribution of the improvement of frying colour after reconditioning, measured as the difference between trait values measured after cold storage at 4 °C and reconditioning. The black bars represent the difference between 4 °C storage and reconditioning. The letters C and E above the bars represent the parental values
The results above represent an independent analysis the starch traits and the traits related to chipping colour. The combination of these two data sets, allow putative relations between starch and chipping quality traits to be determined. The majority of these correlations shown in Table 2 are non-significant, however, with some notable exceptions. A significant correlation (ρ = 0.36, p < 0.001) exists between granule size and cold induced sweetening. Another significant correlation can be observed between chip colour before and after cold storage and starch content (ρ = 0.35, p < 0.001). We conclude that most of the starch related traits, including chipping colour and cold induced sweetening are interconnected.
QTL analysis
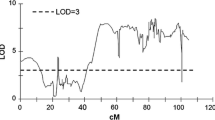
All traits were analysed separately for both years with the exception of the chipping and cold induced sweetening related traits, as no obvious differences were observed between the QTLs for 1998 and 1999. QTLs were observed on eight of the 12 chromosomes using the LOD threshold calculated by permutation test, an overview of the QTL distribution over the map is given in Fig. 4. All QTLs were named according to the trait, the year and the chromosome on which they occur. An overview of all the QTLs and their characteristics is given in Table 3.
QTL analysis of starch phosphorylation revealed 3 QTLs observed in both of the years. The QTLs SP02-2, SP02-5 and SP02-9 explain 21.4, 15.1 and 19.6 % of the phenotypic variance. The corresponding QTLs observed in the other year, explain a larger proportion of the phenotype variance of this trait, (30.3, 29.0 and 18.1 % respectively for SP03-2, SP03-5 and SP03-9). Three QTLs were detected for starch gelling temperature on chromosome 2, 3 and 5. A major QTL indicated as SGT02-2 and SGT03-2 on chromosome 2, explaining 35.8 and 45.5 % of the phenotypic variance, was observed in both years. Two minor QTL STG02-5 and SGT03-3 were significant in only 1 year. For starch granule size, the QTLs on chromosome 8, SGS02-8 and SGS03-8 were consistent in both years, while QTL SGS02-3 is present in 2002 on chromosome 3 and QTL SGS03-9 is present in 2003 on chromosome 9. Amylose content and starch content show the most QTLs of all the starch traits. Five chromosomal regions (2, 3, 5, 7 and 10) are associated with amylose content. The QTLs on chromosomes 2 and 3 were significant in both 2002 and 2003, while the other three QTL were significant in only 1 year. In total five chromosomes (5, 8, 9, 10 and 12) display significant QTL for starch content, and all QTL are consistent in both years. No significant QTL could be identified for enthalpy in either of the 2 years.
QTL analysis of the chipping quality and cold induced sweetening resulted in the identification of five chromosomal regions (3, 5, 8, 9 and 10) influencing chipping colour. Chipping colour directly after harvest resulted in two QTLs on chromosomes 9 and 10. After 3 months storage at 4 °C and after storage and reconditioning, four QTLs are observed on chromosomes 3, 5. 8 and 10 for chipping after storage and 2 QTLs are found for chipping after reconditioning at chromosomes 5 and 10. The explained variances for chipping colour are considerably lower than most of the starch trait QTLs. For cold induced sweetening, three QTLs are detected. Two QTLs on chromosomes 5 and 8, and one QTL at chromosome 5.
Co-localizing QTLs between different years and traits
The QTLs observed in this study are found on 8 of the 12 chromosomes of potato. Within these chromosomes the QTLs of different years or traits overlap as shown in Fig. 4. All QTLs involved in starch content are consistent between years, which is in agreement with the very high phenotypic correlation between years (ρ = 0.72). Equally strong correlations between years were observed for starch phosphorylation and starch gelling temperature, however, the QTLs for the different years of these traits do not always co-localize. For starch phosphorylation this is the case for two of the three QTLs, on chromosomes 5 and 9 and for gelling temperature only one QTL, on chromosome 2, co-localized between years.
On chromosomes 2, 3, 5, 8, 9 and 10 QTLs for multiple traits co-localize, indicating that at these locations there are possible genetic factors that influence multiple starch traits. On chromosome 2, QTLs for degree of phosphorylation, gelling temperature and amylose content overlap. In this case, with the exception of starch phosphorylation, the QTLs for the individual traits are also observed in both years, indicating that starch traits are hardly influenced by environmental variation. At the top of chromosome 3, coinciding QTLs are observed between chipping colour (4 °C) and gelling temperature and at the distal end of the chromosome QTLs coincide for granule size and amylose content. On chromosome 5 there is a large group of overlapping QTLs. In this group we find QTLs for starch phosphorylation, gelling temperature and amylose content, but also QTLs for chipping colour at various time points and cold induced sweetening. On this chromosomal region, both starch characteristics, chipping colour and cold induced sweetening traits cluster. Starch content overlaps on the top part of chromosome 8 with cold induced sweetening and chipping colour. On chromosome 9 QTL involved in starch phosphorylation, chipping colour and granule size cluster. The last cluster, on chromosome 10, contains QTLs for starch content, for chipping colour and amylose content at all three time points. From all these co-localizing QTLs for different starch related traits it is clear that the same genes in the starch biosynthesis are involved in various starch and chipping quality phenotypes.
Candidate gene analysis
Fourteen potato genes involved in carbohydrate metabolism have been mapped in the C × E population. After QTL analysis several of the candidate genes were found to co-localize with QTLs for the starch characteristics, chipping colour and cold induced sweetening. An overview of the candidate genes that map together with the different QTLs is given in Table 3, and their co-localizations are shown in Fig. 4. Genes SSSII, SSSIII, StPho1a, StPho2, GWD and StLin8 co-localise with two or even three different starch related traits.
Discussion
Genetic variation for starch properties
In this study, we analysed the genetic variability of starch content in tubers and five important properties of potato starch, namely starch phosphate content, gelling temperature, enthalpy, granule size and amylose content. These starch properties are important for the various applications of starch as gelling agent in several industrial sectors, including food, textile and construction (Ellis et al. 1998). Chemical and physical (rheological) analysis of starches have been reviewed by Singh et al. (2003), but the wide-ranging genetic variability for starch properties caused by the enzymes involved in granule synthesis and degradation, as well as the trait correlations add to our understanding this versatile polymer. The identification of the chromosomal position of a large number of QTLs for different related starch properties, including the co-localisation with loci encoding enzymes involved in starch metabolism, indicate a refined network of interactions involved in starch synthesis. Subsequently, we aimed to understand chipping quality traits as a function of starch degradation into reducing sugars. In total 38 QTLs are observed over different years for all traits. When this number is corrected for co-segregating QTLs, we end up with 13 QTL locations over eight different chromosomes. Our QTL positions for various starch properties cannot be compared with literature, as these QTLs were not studied previously.
The QTL positions for chipping colour in this study correlate well with the QTL positions for chipping colour as identified by Douches and Freyre (Douches and Freyre 1994) as do the QTL positions for sugar content and cold sweetening as identified by Menendez et al. (Menendez et al. 2002). The strong correlation between reducing sugar content and chipping colour has previously been demonstrated (Scheffler et al. 1992). QTLs for chipping colour in our study confirm that major QTL are present in regions on chromosomes 3, 5, 8, 9 and 10 (Douches and Freyre 1994; Menendez et al. 2002; Bradshaw et al. 2008; Li et al. 2005), although strict comparisons with published data cannot be made due to the lack of shared markers and map resolution. For the chipping QTLs on chromosomes 3 we confirm the co-localisation with starch phosphorylase StPho1a, but the invertase locus PAIN-1 and SSS-1 have been implicated as well (Li et al. 2008). For the QTLs on 5 we propose GWD, while Sut2 proposed by Menendez et al. (Menendez et al. 2002) is also an important candidate. For the QTL on 9 and 10 the apoplastic invertases were proposed (Menendez et al. 2002). QTLs for frying colour, after 4 and 10 °C storage have also been identified (Bradshaw et al. 2008). These QTLs are located on chromosomes 1, 6 and 11. However, these QTLs are not observed in our analysis with the exception of the QTL on chromosome 1 with AGPasS as candidate gene, in the other report on cold induced sweetening (Menendez et al. 2002).
The putative role of candidate genes on starch properties and chipping quality
On chromosome 5 we mapped GWD, this map position is exactly co-localizing with QTLs for starch phosphorylation and cold induced sweetening. GWD is a clear candidate for cold induced sweetening as it stimulates the breakdown of starch by amylases (Edner et al. 2007) and is previously reported to have an effect on cold induced sweetening via the amount of phosphorus it integrates in the starch granules, in this way affecting the degradability of the granules (Lorberth et al. 1998). The main function of GWD is starch phosphorylation by a dikinase-type reaction (Mikkelsen et al. 2004), making it a strong candidate gene for starch phosphorylation as well. Next to that, GWD is also just in the 2LOD interval of the QTLs for starch content, chipping, amylose content and starch gelling temperature. Based on function, approximate position and previous results (Schafer-Pregl et al. 1998) and the fact that on chromosome 5 a co-localization is reported of the QTL for sucrose and fructose content with Sut2 (Barker et al. 2000), Sut2 is probably the most promising candidate gene for at least the starch content QTLs. Marker development in our research for Sut2, on the basis of known exon sequence information did not result in a workable CAPS marker, therefore we were not able to confirm this hypothesis in our research. However the earlier reports (Schafer-Pregl et al. 1998) linked QTLs for starch content to the position of Sut2 gene, suggesting the possibility of two independent QTLs on the upper arm of chromosome 5. For the QTL on chromosomes 9, no co-localizing QTLs are observed in the C × E population. In previous reports however, apoplastic invertases were linked as candidate genes to QTLs on this chromosome (Menendez et al. 2002; Li et al. 2005). On chromosome 9 the invertases were mapped on the upper arm, whereas in our experiments we mapped them on the lower arm of the chromosome. Either the different genetic background or the difference in mapping position could have caused this absence of co-localization. Recently the lack of cold induciblity of the four cell wall invertases on chromosome 9 and 10 was shown, making these genes unlikely candidates (Liu et al. 2010). Furthermore, the link between invertase and chipping colour was explained by the PAIN-1 locus (synonymous to StvacINV1) on chromosome 3 and other members of invertase inhibitor gene family at several chromosomal positions (Liu et al. 2010). Preliminary QTL analysis on an earlier version of our genetic map, without the majority of the candidate genes, did not show QTLs on previously reported positions, such as on chromosome 7. The co-localization of the genes Sps and Sus3 with QTLs for sugar content, however, was reported (Menendez et al. 2002). Our data clearly shows that the QTLs on chromosome 7 were absent; it was thus decided not to develop working markers for these genes. For the same reason, no markers were mapped for Sut1 on chromosome 11.
Most of the reproducible and co-segregating QTL effects in the C × E population are tightly linked to candidate genes at indistinguishable loci. On chromosome 2, QTLs for degree of phosphorylation, gelling temperature and amylose content overlap. For nearly all traits, the QTLs for the different years overlap, indicating a consistent genetic factor regardless of the environment. The LOD profiles of these QTLs co-localize with the map position of SSSII, implying that SSSII is a candidate gene for these traits. SSSII is, together with SSSIII, responsible for the synthesis of amylopectin in the tuber (Edwards et al. 1995). Up to 80 % of the starch granule comprises amylopectin making it the most important constituent of starch. As a consequence, changes in the amylopectin production by SSSII, will most likely have an effect on the total amount of starch produced therefore making a relative increase in amylose content possible by a unaltered amylose production. This explains the match of QTLs Am02-2 and Am03-2 with SSSII even though SSSII is not directly involved in the production of amylose itself. On the contrary, GBSSI, located on chromosome 8, is responsible for amylose production (van de Wal et al. 2001). An explanation for the fact that we do not observe a QTL on chromosome 8 could be due to the fact that the alleles for GBSSI present in the population appear not show a difference in amylose production. Consequently, no QTL is detected at the GBSS locus and QTLs at the positions of other less obvious candidate genes involved in amylose biosynthesis (such as SSSII) co-localise with QTL on chromosome 2. An earlier article describing antisense plants with reduced activity of SSSII (Edwards et al. 1999) an effect of the reduced SSSII expression on the gelatinisation behaviour of starch was found. This is consistent with the co-localization of the QTLs SGT02-2 and SGT03-2 with SSSII. Effects on the gelatinisation behaviour of starch, caused by differences in numbers and lengths of double helices of amylopectin (Cooke and Gidley 1992; Moates et al. 1997) are a good indicator for differences in structure possibly caused by allelic differences in SSSII expression. For starch phosphorylation we find QTLs co-localizing in 1 year with SSSII and in the other year with SSSIII. A clear effect of starch synthases on the degree of phosphorylation is apparent, but there is also an obvious environmental effect shifting the focus between years/environments between both SSSII and SSSIII. Antisense potato lines for SSSII, SSSIII and combined SSSII/SSSIII show clear reductions in phosphorylation levels (Jobling et al. 2002) indicating that both starch synthases have an effect. This effect is possibly caused by the decrease of the amylopectin chain length, reducing the surface to which phosphorus can be attached.
Other genes that appear as candidate genes are two types of starch phosphorylase, more specifically plastidic phosphorylase A (StPho1a) and cytosolic phosphorylase (StPho2). Both genes are involved in a reversible reaction where a glucose-1-phosphate (G1P) is used as substrate in order to add glucose to the α-glucan chain with the release of phosphorus (Pi). StPho1a, in the C × E population, is a candidate gene for onset of starch gelling, chipping and starch granule size. Starch gelling temperature is shown to be affected by the action of Pho1 genes in Pho1 deficient rice plants. It was clearly shown that the onset temperature of starch was significantly lower in the plants with Pho1 deficiency (Satoh et al. 2008). Apart from the change in starch gelling temperature in the same rice experiments, a difference in granule size was observed. The Pho1 deficient plants showed smaller starch granules than the wild type plants. This is in line with the results we obtained. Both StPho1a and StPho2 co-localize with QTLs for starch granule size (SGS03-2 and SGS03-9). Furthermore StPho2 maps together with SP03-9, a QTL for starch phosphorylation. We suggest that this may be because of the action of StPho2, by releasing a Pi, it not only reduces chain length but on the larger scale of the whole granule influences the amount of phosphorus that is incorporated in the granule. Lastly, QTL co-segregating with StPho1a is chipping colour, although for a different time point, a QTL for chipping colour is also observed at the location of StPho2. An explanation for this result can be that StPho2 is involved in degradation of starch as it is hypothesised that, although its actions are reversible, phosphorylases favour a role in the degradation of starch (Satoh et al. 2008).
Conclusions
Combined with the already existing knowledge on genes and interactions within the starch biosynthesis and degradation complex, the results of the QTL analysis in this study provide an increasing insight in the genetic components in potato involved in starch traits and chipping colour. The candidate genes found here, are however so conserved that these results can also be of importance in starch related traits in other crop species. The observed marker-trait correlations, like the link between GWD, starch phosphorylation and chipping quality as trade-off suggests that marker assisted selection for specific GWD alleles allow either for the selection for higher starch phosphate levels or improved chipping quality. In general, these results provide a starting point for further investigations and for indirect selection via molecular markers in an effort to improve the existing potato varieties on the traits related to starch.
References
Barker L, Kuhn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB (2000) SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12(7):1153–1164
Bradshaw JE, Hackett CA, Pande B, Waugh R, Bryan GJ (2008) QTL mapping of yield, agronomic and quality traits in tetraploid potato (Solanum tuberosum subsp tuberosum). Theor Appl Genet 116(2):193–211
Burton WG (1969) The sugar balance in some British potato varieties during storage. II The effects of tuber age, previous storage temperature, and intermittent refrigeration upon low-temperature sweetening. Eur Potato J 12:81–95
Cooke D, Gidley MJ (1992) Loss of crystalline and molecular order during starch gelatinization: origin of the enthalpic transition. Carbohydr Res 227:103–112
Douches DS, Freyre R (1994) Identification of genetic-factors influencing chip color in diploid potato (Solanum Spp). Am Potato J 71(9):581–590
Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan F, Guy C, Smith SM, Steup M, Ritte G (2007) Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol 145(1):17–28
Edwards A, Marshall J, Sidebottom C, Visser RG, Smith AM, Martin C (1995) Biochemical and molecular characterization of a novel starch synthase from potato tubers. Plant J 8(2):283–294
Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rossner U, Martin C, Smith AM (1999) A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J 17(3):251–261
Ellis RP, Cochrane MP, Dale MFB, Duffus CM, Lynn A, Morrison IM, Prentice RDM, Swanston JS, Tiller SA (1998) Starch production and industrial use. J Sci Food Agric 77:289–311
Feingold S, Lloyd J, Norero N, Bonierbale M, Lorenzen J (2005) Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik
Hannemann RE, Peloquin SJ (1967) Crossability of 24-chromosome potato hybrids with 48-chromosome cultivars. Eur Potato J 10:62–73
Hofvander P, Andersson M, Larsson CT, Larsson H (2004) Field performance and starch characteristics of high-amylose potatoes obtained by antisense gene targeting of two branching enzymes. Plant Biotechnol J 2(4):311–320
Hovenkamp-Hermelink JHM, De Vries JN, Adamse P, Jacobsen E, Witholt B, Feenstra WJ (1988) Rapid estimation of the amylose amylopectin ratio in small amounts of tuber and leaf tissue of the potato. Potato Res 31(2):241–246
Jacobsen E (1980) Increase of diplandroid formation and seed set in 4x × 2x crosses in potatoes by genetical manipulation of diphaploids and some theoretical consequences. Z Planzenzuecht 85:110–121
Ji Q, Vincken JP, Suurs LC, Visser RG (2003) Microbial starch-binding domains as a tool for targeting proteins to granules during starch biosynthesis. Plant Mol Biol 51(5):789–801
Jobling SA, Westcott RJ, Tayal A, Jeffcoat R, Schwall GP (2002) Production of a freeze-thaw-stable potato starch by antisense inhibition of three starch synthase genes. Nat Biotechnol 20(3):295–299
Kortstee AJ, Suurs LCJM, Vermeesch AMG, Keetels CJAM, Jacobsen E, Visser RGF (1998) The influence of an increased degree of branching on the physicochemical properties of starch from genetically modified potato. Carbohydr Polym 37(2):173–184
Kossmann J, Lloyd J (2000) Understanding and influencing starch biochemistry. Crit Rev Biochem Mol Biol 35(3):141–196
Kuipers AGJ, Jacobsen E, Visser RGF (1994) Formation and deposition of amylose in the potato-tuber starch granule are affected by the reduction of granule-bound starch synthase gene-expression. Plant Cell 6(1):43–52
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121(1):185–199
Li L, Strahwald J, Hofferbert HR, Lubeck J, Tacke E, Junghans H, Wunder J, Gebhardt C (2005) DNA variation at the invertase locus invGE/GF is associated with tuber quality traits in populations of potato breeding clones. Genetics 170(2):813–821
Li ZH, Lin P, Du SW (2008) Syntheses and characterization of three novel W(Mo)/Cu/S cluster-based layered polymers with 1,3,4-thiadiazole-2,5-dithiolate (SSS): intriguing coordination modes of SSS. Polyhedron 27(1):232–240
Liu X, Song BT, Zhang HL, Li XQ, Xie CH, Liu J (2010) Cloning and molecular characterization of putative invertase inhibitor genes and their possible contributions to cold-induced sweetening of potato tubers. Mol Genet Genomics 284(3):147–159
Lorberth R, Ritte G, Willmitzer L, Kossmann J (1998) Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol 16(5):473–477
Menendez CM, Ritter E, Schafer-Pregl R, Walkemeier B, Kalde A, Salamini F, Gebhardt C (2002) Cold sweetening in diploid potato: mapping quantitative trait loci and candidate genes. Genetics 162(3):1423–1434
Mikkelsen R, Baunsgaard L, Blennow A (2004) Functional characterization of alpha-glucan, water dikinase, the starch phosphorylating enzyme. Biochem J 377(Pt 2):525–532
Moates GK, Noel TR, Parker R, Ring SG (1997) The effect of chain length and solvent interactions on the dissolution of the B-type crystalline polymorph of amylose in water. Carbohydr Res 298(4):327–333
Mullerrober B, Sonnewald U, Willmitzer L (1992) Inhibition of the Adp-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J 11(4):1229–1238
Reid A, Hof L, Felix G, Rucker B, Tams S, Milczynska E, Esselink D, Uenk G, Vosman B, Weitz A (2011) Construction of an integrated microsatellite and key morphological characteristic database of potato varieties on the EU common catalogue. Euphytica 182(2):239–249
Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M (2002) The starch-related R1 protein is an alpha-glucan, water dikinase. Proc Natl Acad Sci USA 99(10):7166–7171
Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang SK, Okita TW, Kaneko N, Fujita N, Yoshida M, Hosaka Y, Sato A, Utsumi Y, Ohdan T, Nakamura Y (2008) Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20(7):1833–1849
Schafer-Pregl R, Ritter E, Concilio L, Hesselbach J, Lovatti L, Walkemeier B, Thelen H, Salamini F, Gebhardt C (1998) Analysis of quantitative trait loci (QTLs) and quantitative trait alleles (QTAs) for potato tuber yield and starch content. Theor Appl Genet 97(5–6):834–846
Scheffler JA, Hesselbach J, Hemme H, Salamini F (1992) Sampling potato genotypes that maintain chip quality under low temperature storage. J Genet Breed 46:103–110
Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi YC, Gidley MJ, Jobling SA (2000) Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat Biotechnol 18(5):551–554
Singh N, Singh J, Kaur L, Sodhi NS, Gill BS (2003) Morphological, thermal and rheological properties of starches from different botanical sources. Food Chem 81(2):219–231
Sowokinos JR (2001) Biochemical and molecular control of cold-induced sweetening in potatoes. Am J Potato Res 78(3):221–236
Stark DM, Timmerman KP, Barry GF, Preiss J, Kishore GM (1992) Regulation of the amount of starch in plant-tissues by Adp glucose pyrophosphorylase. Science 258(5080):287–292
van de Wal MH, Jacobsen E, Visser RG (2001) Multiple allelism as a control mechanism in metabolic pathways: GBSSI allelic composition affects the activity of granule-bound starch synthase I and starch composition in potato. Mol Genet Genomics 265(6):1011–1021
van Eck HJ, Vandervoort JR, Draaistra J, Vanzandvoort P, Vanenckevort E, Segers B, Peleman J, Jacobsen E, Helder J, Bakker J (1995) The inheritance and chromosomal localization of AFLP markers in a noninbred potato offspring. Mol Breed 1(4):397-410
van Ooijen JW (2009) MapQTL® 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen, Netherlands
Visser RGF, Suurs LCJM, Steeneken PAM, Jacobsen E (1997a) Some physicochemical properties of amylose-free potato starch. Starch Starke 49(11):443–448
Visser RGF, Suurs LCJM, Bruinenberg PM, Bleeker I, Jacobsen E (1997b) Comparison between amylose-free and amylose containing potato starches. Starch Starke 49(11):438–443
Acknowledgments
This project was financed by the Centre for BioSystems Genomics (CBSG) which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research. We are grateful to Carolina Celis Gamboa (Laboratory of Plant Breeding, Wageningen University and Research Centre, Wageningen, The Netherlands) for the work she put into the chipping colour phenotyping.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Werij, J.S., Furrer, H., van Eck, H.J. et al. A limited set of starch related genes explain several interrelated traits in potato. Euphytica 186, 501–516 (2012). https://doi.org/10.1007/s10681-012-0651-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-012-0651-y