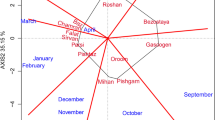
Abstract
Categorization of locations with similar environments helps breeders to efficiently utilize resources and effectively target germplasm. This study was conducted to determine the relationship among winter wheat (Triticum aestivum L.) yield testing locations in South Dakota. Yield trial data containing 14 locations and 38 genotypes from 8 year were analyzed for crossover genotype (G) × environment (E) interactions according to the Azzalini-Cox test. G × E was significant (P < 0.05) and contributed a small proportion of variation over the total phenotypic variation. This suggested that for efficient resource utilization, locations should be clustered. The data were further analyzed using the Shifted Multiplicative Model (SHMM), Spearman’s rank correlation and GGE biplot to group testing locations based on yield. SHMM analysis revealed four major cluster groups in which the first and third had three locations, with four locations in each of the second and fourth groups. Spearman rank correlations between locations within groups were significant and positive. GGE biplot analysis revealed two major mega-environments of winter wheat testing locations in South Dakota. Oelrichs was the best testing location and XH1888 was the highest yielding genotype. SHMM, rank correlation and GGE biplot analyses showed that the locations of Martin and Winner in the second group and Highmore, Oelrichs and Wall in the third group were similar. This indicated that the number of testing locations could be reduced without much loss of grain yield information. GGE biplot provided additional information on the performance of entries and locations. SHMM clustered locations with reduced cross-over interaction of genotype × location. The combined methods used in this study provided valuable information on categorization of locations with similar environments for efficient resource allocation. This information should facilitate efficient targeting of breeding and testing efforts, especially in large breeding programs.





Similar content being viewed by others
References
Abdalla OS, Crossa J, Cornelius PL (1997) Results and biological interpretation of shifted multiplicative model clustering of durum cultivars and test site. Crop Sci 37:88–97
Abou-El-Fittouh HA, Rawlings JO, Miller PA (1969) Classification of environments to control genotype by environment interactions with an application to cotton. Crop Sci 9:135–140
Azzalini A, Cox DR (1984) Two new tests associated with analysis of variance. J R Statist Soc B 46:335–343
Baker RJ (1988) Tests for crossover genotype X environmental interactions. Can J Plant Sci 68:405–410
Byth DE, Eisemann RL, De Lacy IH (1976) Two-way pattern analysis of a large data set to evaluate genotypic adaptation. Heredity 37:215–230
Campbell LG, Lafever HN (1977) Cultivar X environmental interactions in soft red winter wheat yield tests. Crop Sci 17:604–608
Cornelius PL, Seyedsadr M, Crossa J (1992) Using the shifted multiplicative model to search for separability in crop cultivar trials. Theor Appl Genet 84:161–172
Cornelius PL, Van Sanford DA, Seyedsadr M (1993) Clustering cultivars into groups without rank-change interactions. Crop Sci 33:1193–1200
Crossa J, Cornelius PL, Seyedsadr M, Byrne P (1993) A shifted multiplicative model cluster analysis for grouping environments without genotypic rank change. Theor Appl Genet 85:577–586
Fisher RA (1925) Statistical methods for research workers. Oliver and Boyd, Edinburgh, London
Gabriel KR (1971) The biplot graphic display of matrices with application to principal component analysis. Biometrika 58:453–467
Ghaderi A, Everson EH, Cress CE (1980) Classification of environments and genotypes. Crop Sci 20:707–710
Guitard AA (1960) The use of diallel correlations for determining the relative locational performance of varieties of barley. Can J Plant Sci 40:645–651
Horner TW, Frey KJ (1957) Methods for determining natural areas for oat varietal recommendations. Agron J 49:313–315
Jones GL, Matzinger JF, Collins WK (1960) A comparison of flue-cured tobacco varieties repeated over locations and years with implications on optimum plot allocations. Agron J 52:195–199
Lillemo M, van Ginkel M, Trethowan RM et al (2004) Associations among International CIMMYT bread wheat yield testing locations in high rainfall areas and their implications for wheat breeding. Crop Sci 44:1163–1169
Miller PA, Williams JC, Robinson HF (1959) Variety X environments interactions and their implications to testing methods. Agron J 51:132–135
Ramey S, Rosielle AA (1983) HASS cluster analysis: a new method of grouping genotypes or environments in plant breeding. Theor Appl Genet 66:131–133
Roozeboom KL, Schapaugh WT, Tuinstra MR et al (2008) Testing wheat in variable environments: genotype, environment, interaction effects, and grouping test locations. Crop Sci 48:317–330
SAS Institute Inc (2008) SAS OnlineDoc, version 9.1. SAS Institute Inc., Cary, North Carolina
Seyedsadr M, Cornelius PL (1992) Shifted multiplicative models for nonadditive two-way tables. Comm Statist B Simul Comput 21:807–832
Yan W (2001) GGEBiplot-A windows application for graphical analysis of multi-environment trial data and other types of two-way data. Agron J 93:1111–1118
Yan W, Kang MS (2003) GGE biplot analysis-A graphical tool for breeders, geneticists and agronomists. CRC Press, Florida, pp 207–228
Yan W, Hunt LA, Sheng Q et al (2000) Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci 40:597–605
Yau SK, Ferrara GO, Srivastava JP (1991) Classification of diverse bread-wheat growing environments based on differential yield responses. Crop Sci 31:571–576
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Malla, S., Ibrahim, A.M.H., Little, R. et al. Comparison of shifted multiplicative model, rank correlation, and biplot analysis for clustering winter wheat production environments. Euphytica 174, 357–370 (2010). https://doi.org/10.1007/s10681-010-0130-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0130-2