Summary
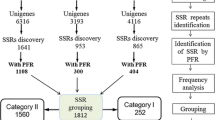
Comparative genetic analysis has shown that different plant species often share orthologous genes for very similar functions, especially for cereal crops. In this study, we focused on analyzing the utility of EST-SSR markers among monocots in silico and by experiment. Based on 423,611 wheat ESTs, 101,299 were found to express commonly in rice, maize and barley ESTs, which accounted for 23.94% of wheat EST data. 1707 SSR-containing ESTs (SSR-ESTs) were mined from the 101,299 homologous ESTs, which accounted for 8.8% of all the 19,434 wheat EST-SSRs. It showed that the number of homologous SSR-ESTs much less than the homologous ESTs among grasses. The number of conserved ESTs and SSR-ESTs were inversely proportional to the evolutionary distance among species. Although these SSR-ESTs were in high similarity, the number of conserved SSRs between wheat and the other three crops decreased with the increased aligning regions, and only 13 types of SSR motifs in 21 ESTs were obtained by aligning sequences over a stretch of more than 40 bp. The variation at a microsatellite came from not only the differences in the numbers of repeat units and its location shift, but also base mutations flanking microsatellite repeats. From the 428 uni-SSR-ESTs universally in the four cereals, 243 primers were designed and tested in two genotypes of each species. 154 (63.4%) primers produced clear amplicons across the four species, which showed a high efficient transferability of wheat EST-SSR markers to the other three cereal crops.
Similar content being viewed by others
References
Ahn, S. & S.D. Tanksley, 1993. Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA 90: 7980–7997.
Van Deynze, A.E., M.E. Sorrells, W.D. Park, N.M. Ayres, H. Fu, S.W. Cartinhour, E. Paul & S.R. McCouch, 1998. Anchor Probes for Comparative Mapping of Grass genera. Theor Appl Genet 97: 356–369.
Altschul S.F., T.L. Madden, A.A.Schaffer, J Zhang, Z Zhang, W Miller and D.J.Lipman, 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Bennetzen, J.L. & W. Ramakrishna, 2002. Numerous small rearrangements of gene content, order, and orientation differentiate grass genomes. Plant Mol Biol 48: 821–827.
Cordeiro, G.M., R. Casu, C.L. McIntyre, J.M. Manners & R.J. Henry, 2001. Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross-transferable to erianthus and sorghum. Plant Sci 160: 1115–1123.
Decroocq, V., M.G. Fave, L. Hagen, L. Bordenave & S. Decroocq, 2003. Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106: 912–922.
Devos, K.M. & M.D. Gale, 2000. Genome relationships: the grass model in current research. Plant Cell 12: 637–646.
Devos, K.S., S. Chao, Q.Y. Li, M.C. Simonetti & M.D. Gale, 1994. Relationship between chromosome 9 of maize and wheat homologous group 7 chromosomes. Genetics 138: 1287–1292.
Eujayl, I., M.E. Sorrells, M. Baum, P. Wolters & W. Powell, 2001. Assessment of genotypic variation among cultivated durum wheat based on EST-SSRs and genomic SSRs. Euphytica 119: 39–43.
Eujayl, I., M.E. Sorrells, M. Baum, P. Wolters & W. Powell, 2002. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet 104: 399–407.
Fatokun, C.A., D.I. Menacio-Hautea, D. Danesh & N.D. Young, 1992. Evidence for orthologous seed weight genes in cowpea and mungbean based upon RFLP mapping. Genetics 132: 841–846.
Gao, L., J. Tang, H. Li & J. Jia, 2003. Analysis of microsatellites in major crops assessed by computational and experimental approaches. Mol Breed 12: 245–261.
Gao, L.F., R.L. Jing, N.X. Huo, Y. Li, X.P. Li, R.H. Zhou, X.P. Chang, J.F. Tang, Z.Y. Ma & J.Z. Jia, 2004. One hundred and one new microsatellite loci derived from ESTs (EST-SSRs) in bread wheat. Theor Appl Genet 108: 1392–1400.
Gaut, B.S., 2002. Evolutionary dynamics of grass genomes. New Phytol 154: 15–28.
Gupta, P.K., S. Rustgi, S. Sharma, R. Singh, N. Kumar & H.S. Balyan, 2003. Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genomics 270: 315–323.
Kantety, R.V., M.L. Rota, D.E. Matthews & M.S. Sorrells, 2002. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48: 501–510.
La Rota, M. & M.E. Sorrells, 2004. Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Funct Integr Genomics 4: 34–46.
Leigh, F, V. Lea, J. Law, P. Wolters, W. Powell & P. Donini, (2003) Assessment of EST- and genomic microsatellite markers for variety discrimination and genetic diversity studies in wheat. Euphytica 133: 359–366.
Li, Y.C., A.B. Korol, T. Fahima & E. Nevo, 2004. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 21: 991–1007.
Matsuoka, Y., S.E. Mitchell, S. Kresovich, M. Goodman & J. Doebley, 2002. Microsatellite in Zea-variability, patterns of mutations, and use for evolutionary studies. Theor Appl Genet 104: 436–450.
Moore, G., K.M. Devos, Z. Wang & M.D. Gale, 1995. Grasses, line up and form a circle. Curr Biol 5:737–739.
Sorrells, ME. et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13: 1817–1827.
Taylor, C., K. Madsen, S. Borg, M.G. Moller, B. Boelt & P.B. Holm, 2001. The development of sequence-tagged sites (STSs) in Lolium perenne L.: The application of primer sets derived from other genera. Theor Appl Genet 103: 648–658.
Varshney, R.K., R. Sigmund, A. Borner, V. Korzun, N. Stein, M.E. Sorrells, P. Langridge & A. Graner, 2005. Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci 168: 195–202.
Yu, J.K., T.M. Dake, S. Singh, D. Benscher, W. Li, B. Gill & M.E. Sorrells, 2004a. Development and mapping of EST-derived simple sequence repeat (SSR) markers for hexaploid wheat. Genome 47: 805–818.
Yu, J.K., M. La Rota, R.V. Kantety & M.E. Sorrells, 2004b. EST derived SSR markers for comparative mapping in wheat and rice. Mol Gen Genomics 271: 742–751.
Zhang, L.Y., M. Bernard, P. Leroy, C. Feuillet & P. Sourdille, 2005. High transferability of bread wheat EST-derived SSRs to other cereals. Theor Appl Genet 111: 677–687.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tang, J., Gao, L., Cao, Y. et al. Homologous analysis of SSR-ESTs and transferability of wheat SSR-EST markers across barley, rice and maize. Euphytica 151, 87–93 (2006). https://doi.org/10.1007/s10681-006-9131-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-006-9131-6