Abstract
Effective surveillance for epidemic-prone viral diseases is essential for emergency preparedness to respond to threats and occurrences of pandemics. While it is difficult and expensive to conduct health facility-based surveillance, there is a growing interest in conducting sewage-based epidemiological studies to monitor the health of the urban population because of the relative ease of sample collection and the availability of advanced molecular techniques for the detection of pathogens in the sewage. Sewage samples offer unique means to study the aggregate health of the population as opposed to the monitoring of the health of any individual by traditional methods. We worked together with the Ministry of Public Works in Kuwait and developed a platform for the collection and testing of sewage samples from different regions of Kuwait for studying population health. In this report, we describe the results of a cross-sectional study conducted between 16 and 23 September 2019 in an attempt to detect influenza, Noro, Rota, hepatitis A, and hepatitis E viruses in urban sewage samples collected in Kuwait. All five targeted viruses were detected in the samples collected from urban wastewater in Kuwait using reverse-transcriptase quantitative PCR (RT-qPCR). We recently checked for the presence of SARS-CoV-2 in the stored cDNA samples and confirmed the absence of SARS-CoV-2 in them. This is the first report that demonstrates the preparedness in Kuwait for using sewage samples for the detection and monitoring of many pathogenic viruses which may greatly increase the capacity of the country to deal with a viral disease outbreak in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The world is finally recovering from the worst disease pandemic in human history that recorded a death toll of over 6.4 million due to COVID-19 (WHO, 2022). Infectious disease surveillance remains the cornerstone for early detection and control of highly contagious diseases in humans. The use of molecular techniques like real-time polymerase chain reaction (PCR) ushered in a new era of rapid diagnosis of infectious diseases enabling more effective surveillance of epidemic-prone viral diseases like influenza. Reliable and rapid diagnostic tools have thus become essential for emergency preparedness to respond to threats and occurrences of pandemic influenza. Health facility-based surveillance remains the norm in most countries for monitoring infectious disease outbreaks even though it does not cover the disease activity in the whole community. Even during a typical outbreak investigation, limited information may be obtained by tracing the suspected individuals and their contacts. Traditional surveillance system for infectious diseases fails to detect asymptomatic individuals who may continue to spread the disease in the community. Thus, there is a need for novel ways to monitor large groups of people without testing every healthy person in the community. Urban areas that are equipped with modern sewerage system offer unique opportunities to monitor the health of the population without individual testing since wastewater provides an aggregate sample from the whole community (Carey, 2019). Some viruses are shed in the stool and urine of the infected persons and may be detected for a variable period in the sewage with the help of powerful molecular techniques (Majumdar et al., 2018a). Kuwait has a highly developed sewage network and sewage treatment facilities that include six wastewater treatment plants across the country located at Al Jahra, Sulaibiya, Kabd, Al Riqqa, Ardhiya, and Umm Al Hayman areas (Kuwait Environmental Public Authority, 2016). Ninety percent of the Kuwaiti population has access to water and sanitation services as they live in the urban areas (Fanack Water, 2020). Thus, sewage-based epidemiology may significantly help address many public health issues in this country including the monitoring of infectious disease outbreaks. We selected five viruses, namely influenza, Noro, Rota, hepatitis A, and hepatitis E viruses, for their detection in the wastewater of Kuwait as they have the potential for an outbreak and are shed in the feces.
Influenza is an infectious respiratory disease mainly caused by influenza A and influenza B viruses in humans (Krammer et al., 2018). The symptoms of influenza may be mild in the majority with a sore throat, fever, runny nose, cough, headache, and muscle pain for about 1 week with an uneventful recovery. Some patients have more severe symptoms due to viral pneumonia requiring hospitalization and may even die from respiratory failure. The influenza pandemic of 1918–1919 which killed at least 20 million people and the pandemics in 1957 and 1968 underscored the importance of making preparations to face future pandemics, and having surveillance programs to monitor the re-emergence of more virulent forms of the virus (Institute of Medicine, 2005).
Norovirus is the leading cause of acute gastroenteritis globally (Kirk et al., 2015). Norovirus infection occurs in all age groups but predominantly affects the young children. Diarrhea and vomiting are the major clinical symptoms of norovirus infection which may be accompanied by abdominal cramps and fever. Most infections result in full recovery within 3 days, but more severe illnesses, sometimes leading to death, may occur in young children, elderly over 65 years of age, and immunocompromised hosts. Rotavirus infections mainly affect children less than 5 years of age causing severe, dehydrating gastroenteritis. In spite of the global vaccination efforts, more than 200,000 deaths result from rotavirus infections annually, mostly occurring in low-income countries (Crawford et al., 2017).
Epidemics due to viral hepatitis dates back to ancient times; however, there has been a significant decline in the incidence of this deadly disease with technological advancements. Hepatitis A and E viruses cause acute, self-limited inflammatory disease of the liver (Lemon et al., 2019). Eighty percent of adults infected with hepatitis A develop severe hepatitis with jaundice, but in children, less than 30% of cases develop symptomatic hepatitis. Hepatitis E is the most common cause of acute viral hepatitis worldwide. However, it follows a more benign course in most people except in some vulnerable groups including pregnant women, older people, and immunocompromised hosts who may have a serious outcome. Both hepatitis A and E viruses share a common fecal–oral transmission and spread by the consumption of contaminated food or water and person-to-person contact.
Kuwait is inhabited by over 4.1 million people all of whom reside in urban areas (The World Bank, 2018). The municipal sewage network covers all the six governorates of Kuwait, namely Al-Asimah, Jahra, Hawally, Farwaniya, Mubarak Al-Kabeer, and Al-Ahmadi. Thus, all the inhabitants of Kuwait have access to a modern sewerage system which is maintained by the Ministry of Public Works (MPW). The MPW performs routine analysis of treated wastewater which includes counting of coliforms as the only microbiological assessment. It was hypothesized that common pathogenic viruses with outbreak potentials could be detected by analyzing untreated wastewater collected in the community by using molecular techniques. A cross-sectional study was conducted in Kuwait between 16 and 23 September 2019 in an attempt to detect influenza, Noro, Rota, hepatitis A, and hepatitis E viruses in urban sewage samples collected from all the governorates of Kuwait.
Methods
Sample collection
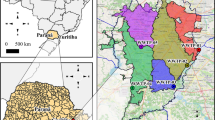
Six sampling sites were selected randomly one from each governorate of Kuwait, namely Jaber Al Ali, Jabriya, Waha, Adan, Ardiya, and Qurtuba while ensuring a population size of at least 5000 in each of the selected catchment areas by using data from Public Authority for Civil Information (Fig. 1). One liter of sewage sample was collected using a sterile glass bottle from a manhole of each of the selected locations between September 16 and 23, 2019.
Extraction of RNA
The samples were prepared for RNA isolation according to Laverick et al. (2004) with minor modification to optimize the method for the detection of the viruses in a smaller volume of sewage. Beef extract solution was added to the sewage sample after initial filtration with a filter paper to remove the large debris. The sample was centrifuged at 7000 × g at 4 °C for 30 min, and the supernatant was flocculated at pH 3.5. Then, the sample was centrifuged again for 30 min, and the floc was dissolved in the buffer. Viral RNA was extracted from the dissolved floc using QIAamp Viral RNA Mini Kit. Reverse transcription was performed to create cDNA from the RNA, and then PCR was performed using the transcribed cDNAs and the specific primers of the targeted viruses. Purified RNA prepared from plasmids containing the conserved domains was used as controls during the gel electrophoresis.
Reverse transcriptase gel-based qualitative PCR
Reverse transcription
After viral RNA extraction, reverse transcription was carried out using iScript Supermix (Bio-Rad) to generate cDNA. For these reactions, 2 µl of the RNA template was mixed with 4 µl of RT supermix and nuclease-free water to make a total reaction volume of 20 µl. RT was performed under the following reaction conditions: 25 °C for 5 min, 46 °C for 20 min, and then 95 °C for 1 min.
Polymerase chain reaction (PCR)
PCR was performed in a T100 thermal cycler (Bio-Rad Laboratories). For the amplification reactions (25 µl total volume), 1 µl of cDNA was used as a template + 1 µl of each primer (10 µM) + 2.5 µl of dNTPs + 2 µl of MgCl2 + 0.5 µl of Taq polymerase + 2.5 µl of 10 × buffer and distilled water. The reaction conditions were as follows: an initial denaturation stage at 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s, 55 °C for 30 s, 72 °C for 1 min, and finally 72 °C for 10 min.
Gel electrophoresis
The PCR products were run on 1.2% certified megabase agarose gel in 1X Tris/Borate/EDTA (TBE) buffer at 120 V at 90 min for gel electrophoresis. One hundred base pair DNA ladder (NE Biolabs) was used for sizing the bands. Bands were visualized using the ChemiDoc MP imaging system (Bio-Rad Laboratories). All gels were stained with ethidium bromide. The images were processed, and bands were analyzed with Image Lab Software 5.0 (Bio-Rad Laboratories Inc.) to estimate the sizes of the PCR products.
Quantitative real-time PCR (qPCR)
Preparation of in vitro transcribed RNA for qPCR
In vitro transcription was performed using the MEGAscript® Kit following the instructions provided by the manufacturer (MEGAscript manual 1330 M). In vitro transcribed RNA standards were generated from pBluescript (UW5VIR) plasmid DNA (MIT, USA). Subcloning efficiency DH5α competent cells were used for transformation (Invitrogen). pUC19 DNA supplied with the kit was used as a control to verify the transformation efficiency of the cells. Luria–Bertani (LB) medium (Conda) was routinely used to culture E. coli. Antibiotic plates for the selection of transformants contained ampicillin (Sigma–Aldrich) at a final concentration of 100 µg/ml. Plasmid DNA isolated using PureLink HQ Mini Plasmid Purification Kit (Invitrogen) was linearized using KpnI restriction endonuclease (New England Biolabs, UK) according to the manufacturer’s instruction. A total of 2 µg of plasmid DNA (at a concentration of 238.4 ng/ul) was used and digested by adding 10 U of KpnI enzyme in NEbuffer provided by the manufacturer. The digestion mixture was incubated at 37 °C for 3 h followed by thermal inactivation at 80 °C for 20 min. The digested DNA was run in 0.8% certified megabase agarose gel in 1X Tris/Borate/EDTA (TBE) buffer at 60 V at 100 min for agarose gel electrophoresis. The digested DNA was gel purified using the QIA quick Gel Extraction kit (Qiagen). A total of 520 ng/µl was the concentration of DNA measured using IMPLEN-Nanophotometer P-Class. In vitro transcription was carried out with 6 µl of digested plasmid DNA using MEGAscript® T7 Kit (Invitrogen) and purified using QIAamp Viral RNA Mini Kit, according to the manufacturer’s instructions and eluted in a final volume of 40 µl elution buffer. RNA concentration (62 ng/µl) was estimated using the Nanodrop 1000 spectrophotometer (Thermo Scientific, UK).
qPCR
Quantification using qPCR was performed using the CFX96 cycler (Bio-Rad). Standard curves were generated from tenfold serial dilution series of the in vitro transcribed RNA. The reaction mixtures contained the RNA sample + 2X iTaq Universal Probes One-step mix + primers and probe for each of the 5 viruses. RT PCR assay was based on Flu_Fw ARATGAGTCTTCTRACCGAGGTCG, Flu_Rv TGCAAAGACATCYTCAAGYYTCTG and Flu_Pb TCAGGCCCCCTCAAAGCCGA, Noro_Fw TGGAYTTTTAYGTGCCCAG, Noro_Rv CGACGCCATCTTCATTCAC and Noro_Pb AGCCAGATTGCGATCGCCC, HAV_Fw TCACCGCCGTTTGCCCTAG, HAV_Rv GGAGAGCCCTGGAAGAAAG and HAV_Pb CCTGAACCTGCAGGAATTAA, HEV_Fw CGGTGGTTTCTGGGGTGAC, HEV Rv GAAGGGGTTGGTTGGATGAA and HEV_Pb CGGGTTGATTCTCAGCCCTTCGC, Rota_Fw CAGTGGTTGHTGCTCAAGATGGA, Rota_Rv TCATTGTAATCATATTGAATACCCA and Rota _Pb ACAACTGCAGCTTCAAAAGAAGWGT, respectively. Cycling conditions consisted of a reverse transcription step at 50 °C for 10 min, followed by polymerase activation for 2 min at 95 °C and 40 cycles of denaturation for 15 s at 95 °C and annealing/extension for 30 s at 60 °C.
Results and discussion
Wastewater-based epidemiology has dramatically changed the scenario of disease surveillance especially in urban areas equipped with modern sewerage by providing the opportunity for early detection of an infectious disease outbreak due to a known microorganism. In spite of the limitations of detection of many organisms due to their low concentration, powerful molecular techniques have enabled the detection of even RNA viruses like SARS-CoV-2 which prompted worldwide initiatives to detect and monitor the spread of the virus by wastewater sampling (Bivins et al., 2020). The present study was conducted only a few months before the outbreak of SARS-CoV-2 to test the hypothesis if common pathogenic viruses could be detected in the sewage by using modern techniques for the separation of biomolecules (Jamil et al., 2020). Instead of targeting any particular strain, primers were chosen to amplify a conserved domain of all the influenza viruses (Hussein et al., 2016a, b; Runstadler et al., 2013). Primers were also used for the detection of Rota, Noro, hepatitis A, and hepatitis E viruses as they have the potential for causing seasonal or occasional outbreaks (Jothikumar et al., 2006, 2009; Costafreda et al., 2006; Hellmer et al., 2014).
Using specific primers in reverse transcription gel-based PCR, bands of the expected sizes representing all 5 viruses were detected in all six selected sites, except for the hepatitis E virus (HEV) (Figs. 2, 3, and 4). HEV was below the detection limit of traditional PCR in Ardiya and was present at very low levels in Qurtuba. This indicates that these foodborne viruses are widely circulating in Kuwait. In quantitative real-time PCR (qPCR), amplification curves for both standards and samples were detectable, which indicates that the whole workflow starting from sample processing to qPCR detection is functional. All samples were tested in duplicates, and there were no detectable discrepancies between those replicates, which reflects good pipetting techniques. All five viruses were detected by qPCR in samples collected from every location (Table 1). Interestingly, high influenza activity was detected in Jabriya (1012 genome copies). It should be mentioned here that the sampling location in Jabriya was very close to Mubarak Al Kabeer Hospital, one of the largest tertiary hospitals in Kuwait which serves nearly 700,000 people of Hawally Governorate. The high influenza activity in Jabriya may therefore represent an actually high number of influenza cases in that area as compared with lower values observed in the other sampling locations representing residential areas. Surveillance data from WHO Eastern Mediterranean Regional Office shows a variable influenza activity in this region throughout the year (WHO, 2019). In 2019, influenza activity in the region was high in April and gradually declined to low levels in August and rose again in the months of September and October. Due to the high sensitivity of qPCR, we were able to detect HEV using this technique in the samples that were negative by the traditional qualitative PCR. It is noteworthy that the hepatitis E virus was not detectable by RT-PCR in Ardiya and very weakly detectable in Qurtuba although the virus was detected by RT-qPCR in both areas at a lower concentration than the other viruses. It shows the superiority of qPCR over traditional PCR with respect to sensitivity and therefore suggesting the former technique to be more suitable for the detection of targeted microorganisms.
In this study, a group of viruses were detected in the sewage when there was no ongoing outbreak of the same viruses in the community. Serial testing at intervals would allow monitoring of the ongoing disease activity in the community and predict a possible outbreak if increasing levels are detected. SARS-CoV-2 was isolated from wastewater in a city in the Netherlands, only 4 days after the first case of COVID-19 was reported showing the prospects of wastewater analysis as an early warning system for an outbreak (Mallapaty, 2020). Routine surveillance for SARS-CoV-2 in wastewater samples are now being carried out in many countries including the USA, the UK, Canada, Australia, Sweden, and Denmark for monitoring the disease activity in the community. The use of sewage-based epidemiology for tracking pathogenic viruses globally may serve as an effective early warning system and help prevent catastrophic pandemics like COVID-19 in the future.
Sewage-based surveillance can play an important role in identifying the re-emergence of other pathogens that were targeted for eradication from the globe. The use of a cell culture system, PCR, and next-generation sequencing made it possible to detect poliovirus even when its concentration becomes very low. Environmental surveillance has been used successfully by global polio eradication program (Majumdar et al., 2018b). Very recently, the detection of poliovirus in the wastewater in London prompted the health authority to roll out urgent polio vaccination for all London-based children under 10 (Hussain, 2022).
Conclusion
This study documents the detection of five pathogenic viruses in the wastewater in Kuwait using molecular biology techniques. The capacity for detection of influenza, Noro, Rota, hepatitis A, hepatitis E, and SARS-CoV-2 viruses in the sewage greatly enhances the ability of the Ministry of Health of Kuwait to efficiently deal with outbreaks caused by the same viruses in the future. Sewage-based surveillance can also be employed to monitor the re-emergence of other pathogens that are targeted for eradication including the poliovirus.
Data availability
All data related to this study are available on request.
References
Bivins, A., North, D., Ahmad, A., Ahmed, W., Alm, E., Been, F., Bhattacharya, P., Bijlsma, L., Boehm, A. B., Brown, J., Buttiglieri, G., Calabro, V., Carducci, A., Castiglioni, S., Cetecioglu Gurol, Z., Chakraborty, S., Costa, F., Curcio, S., de Los Reyes, F. L., 3rd, Delgado Vela, J., & Bibby, K. (2020). Wastewater-based epidemiology: Global collaborative to maximize contributions in the fight against COVID-19. Environmental Science & Technology, 54(13), 7754–7757. https://doi.org/10.1021/acs.est.0c02388
Carey, J. (2019). News feature: Interested in gauging a population’s health? Look to sewage. Proceedings of the National Academy of Sciences of the United States of America, 116(13), 5836–5839. https://doi.org/10.1073/pnas.1903138116
Costafreda, M. I., Bosch, A., & Pintó, R. M. (2006). Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Applied and Environmental Microbiology, 72(6), 3846–3855. https://doi.org/10.1128/AEM.02660-05
Crawford, S. E., Ramani, S., Tate, J. E., Parashar, U. D., Svensson, L., Hagbom, M., Franco, M. A., Greenberg, H. B., O’Ryan, M., Kang, G., Desselberger, U., & Estes, M. K. (2017). Rotavirus Infection. Nature Reviews Disease Primers, 3, 17083. https://doi.org/10.1038/nrdp.2017.83
Fanack Water (2020). Water infrastructure of Kuwait. Retrieved September 20, 2022, from https://water.fanack.com/kuwait/water-infrastructure-in-kuwait/
Hellmér, M., Paxéus, N., Magnius, L., Enache, L., Arnholm, B., Johansson, A., Bergström, T., & Norder, H. (2014). Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Applied and Environmental Microbiology, 80(21), 6771–6781. https://doi.org/10.1128/AEM.01981-14
Hussain, Z. (2022). Polio vaccine is offered to all children in London aged 1 to 9 after virus detected in sewage. BMJ, 378, o2007. https://doi.org/10.1136/bmj.o2007
Hussein, I. T., Krammer, F., Ma, E., Estrin, M., Viswanathan, K., Stebbins, N. W., Quinlan, D. S., Sasisekharan, R., & Runstadler, J. (2016a). New England harbor seal H3N8 influenza virus retains avian-like receptor specificity. Scientific Reports, 6, 21428. https://doi.org/10.1038/srep21428
Hussein, I., Ma, E. J., Hill, N. J., Meixell, B. W., Lindberg, M., Albrecht, R. A., Bahl, J., & Runstadler, J. A. (2016b). A point mutation in the polymerase protein PB2 allows a reassortant H9N2 influenza isolate of wild-bird origin to replicate in human cells. Infection, Genetics and Evolution : Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 41, 279–288. https://doi.org/10.1016/j.meegid.2016.04.011
Institute of Medicine. (2005). The threat of pandemic influenza: Are we ready? Workshop summary. The National Academies Press.
Jamil, K., Abdulrazack, N., Fakhraldeen, S., Kumar, V., Alsubiai, S., Al-Ati, T., Baron, H., Husain, F. I., Ahmed, I., & Hussein, I. (2020). Detection of pathogenic viruses in the urban wastewater in Kuwait. In 2020 ASM Biothreats; Arlington, Virginia, U.S.A.
Jothikumar, N., Cromeans, T. L., Robertson, B. H., Meng, X. J., & Hill, V. R. (2006). A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. Journal of Virological Methods, 131(1), 65–71. https://doi.org/10.1016/j.jviromet.2005.07.004
Jothikumar, N., Kang, G., & Hill, V. R. (2009). Broadly reactive TaqMan assay for real-time RT-PCR detection of rotavirus in clinical and environmental samples. JIN2@cdc.gov. Journal of Virological Methods, 155(2), 126–131. https://doi.org/10.1016/j.jviromet.2008.09.025
Kirk, M. D., Pires, S. M., Black, R. E., Caipo, M., Crump, J. A., Devleesschauwer, B., Döpfer, D., Fazil, A., Fischer-Walker, C. L., Hald, T., Hall, A. J., Keddy, K. H., Lake, R. J., Lanata, C. F., Torgerson, P. R., Havelaar, A. H., & Angulo, F. J. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Medicine, 12(12), e1001921.
Krammer, F., Smith, G., Fouchier, R., Peiris, M., Kedzierska, K., Doherty, P. C., Palese, P., Shaw, M. L., Treanor, J., Webster, R. G., & García-Sastre, A. (2018). Influenza. Nature Reviews Disease Primers, 4(1), 3.
Kuwait Environment Public Authority. (2016). Kuwait environmental portal. Kuwait waste – waste management (Beatona). Retrieved September 20, 2022, from https://gisportal.emisk.org/arcgis/apps/MapJournal/index.html?appid=e53ce22b1cd148189fec322a828a6419
Laverick, M. A., Wyn-Jones, A. P., & Carter, M. J. (2004). Quantitative RT-PCR for the enumeration of noroviruses (Norwalk-like viruses) in water and sewage. Letters in Applied Microbiology, 39(2), 127–136. https://doi.org/10.1111/j.1472-765X.2004.01534.x
Lemon, S. M., & Walker, & C. M. (2019). Hepatitis A virus and hepatitis E virus: Emerging and re-emerging enterically transmitted hepatitis viruses. Cold Spring Harbor Perspectives in Medicine, 9(6), a031823.
Majumdar, M., Sharif, S., Klapsa, D., Wilton, T., Alam, M. M., Fernandez-Garcia, M. D., Rehman, L., Mujtaba, G., McAllister, G., Harvala, H., Templeton, K., Mee, E. T., Asghar, H., Ndiaye, K., Minor, P. D., & Martin, J. (2018a). Environmental surveillance reveals complex enterovirus circulation patterns in human populations. Open Forum Infectious Diseases, 5(10), ofy250. https://doi.org/10.1093/ofid/ofy250
Majumdar, M., Klapsa, D., Wilton, T., Akello, J., Anscombe, C., Allen, D., Mee, E. T., Minor, P. D., & Martin, J. (2018b). Isolation of vaccine-like poliovirus strains in sewage samples from the United Kingdom. The Journal of Infectious Diseases, 217(8), 1222–1230. https://doi.org/10.1093/infdis/jix667
Mallapaty, S. (2020). How sewage could reveal true scale of coronavirus outbreak. Nature, 580(7802), 176–177. https://doi.org/10.1038/d41586-020-00973-x
Runstadler, J., Hill, N., Hussein, I. T., Puryear, W., & Keogh, M. (2013). Connecting the study of wild influenza with the potential for pandemic disease. Infection, Genetics and Evolution : Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 17, 162–187. https://doi.org/10.1016/j.meegid.2013.02.020
The World Bank. (2018). Retrieved June 24, 2020, from https://data.worldbank.org/indicator/SP.POP.TOTL?locations=KW
World Health Organization. (2019). Retrieved September 19, 2022, from http://www.emro.who.int/pandemic-epidemic-diseases/influenza/influenza-monthly-update-september-2019.html
World Health Organization. (2022). Retrieved September 18, 2022, from https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---14-september-2022
Acknowledgements
The authors gratefully acknowledge collaborative support from Prof Eric Alm, Prof Carlo Ratti, Dr. Fabio Duarte, Dr. Mariana Matus, and Ms. Newsha Ghaeli of Massachusetts Institute of Technology, USA, for method developments for this study. The authors also thank the Ministry of Public Works of Kuwait for technical support during the collection of the study samples.
Funding
The authors received funding from the Kuwait Foundation for Advancement of Sciences (KFAS grant number P315-75EV-01) to conduct this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jamil, K., Abdulrazack, N., Fakhraldeen, S. et al. Detection of pathogenic viruses in the urban wastewater in Kuwait—implications for monitoring viral disease outbreaks. Environ Monit Assess 195, 406 (2023). https://doi.org/10.1007/s10661-023-10986-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-023-10986-4