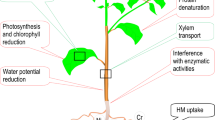
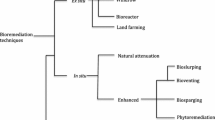
Abstract
Soil heavy metals pollution can cause many serious environment problems because of involving a very complex pollution process for soil health. Therefore, it is very important to explore methods that can effectively evaluate heavy metal pollution. Researchers were actively looking for new ideas and new methods for evaluating and predicting levels of soil heavy metal pollution. The study on microbial communities is one of the effective methods using gene chip technology. Gene chip technology, as a high-throughput metagenomics analysis technique, has been widely used for studying the structure and function of complex microbial communities in different polluted environments from different pollutants, including the soil polluted by heavy metals. However, there is still a lack of a systematic summarization for the polluted soil by heavy metals. This paper systematically analyzed soil heavy metals pollution via reviewing previous studies on applying gene chip technology, including single species, tolerance mechanisms, enrichment mechanisms, anticipation and evaluation of soil remediation, and multi-directional analysis. The latest gene chip technologies and corresponding application cases for discovering critical species and functional genes via analyzing microbial communities and evaluating heavy metal pollution of soil were also introduced in this paper. This article can provide scientific guidance for researchers actively investigating the soil polluted by heavy metals.







Similar content being viewed by others
Change history
28 February 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10661-022-09797-w
References
Assuno, A. G. L., Schat, H., & Aarts, M. G. M. (2003). Thlaspi caerulescens, an attractive model species to study heavy metal hyperaccumulation in plants. New Phytologist. https://doi.org/10.1046/j.1469-8137.2003.00820.x
Azarbad, H., Niklinska, M., Laskowski, R., van Straalen, N. M., van Gestel, C. A., Zhou, J., He, Z., Wen, C., & Roling, W. F. (2015). Microbial community composition and functions are resilient to metal pollution along two forest soil gradients. FEMS Microbiology Ecology, 91(1), 1–11. https://doi.org/10.1093/femsec/fiu003
Azevedo, H., Azinheiro, S. G., Muñoz-Mérida, A., Castro, P. H., Huettel, B., Aarts, M. G., & Assunção, A. G. (2016). Transcriptomic profiling of Arabidopsis gene expression in response to varying micronutrient zinc supply. Genomics Data, 7, 256–258. https://doi.org/10.1016/j.gdata.2016.01.021
Bai, S., Li, J., He, Z., Van Nostrand, J. D., Tian, Y., Lin, G., Zhou, J., & Zheng, T. (2013). GeoChip-based analysis of the functional gene diversity and metabolic potential of soil microbial communities of mangroves. Applied Microbiology and Biotechnology, 97(15), 7035–7048. https://doi.org/10.1007/s00253-012-4496-z
Becher, M., Talke, I. N., Krall, L., & Krämer, U. (2003). Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant Journal, 37(2), 251–268. https://doi.org/10.1046/j.1365-313x.2003.01959.x
Black, A., Wakelin, S., Hamonts, K., Gerard, E., & Condron, L. (2019). Impacts of long term copper exposure on abundance of nitrogen cycling genes and denitrification activity in pasture soils. Applied Soil Ecology, 138, 253–261. https://doi.org/10.1016/j.apsoil.2019.03.009
Brodie, E. L., DeSantis, T. Z., Joyner, D. C., Baek, S. M., Larsen, J. T., Andersen, G. L., Hazen, T. C., Richardson, P. M., Herman, D. J., & Tokunaga, T. K. (2006). Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Applied and Environment Microbiology, 72(9), 6288–6298. https://doi.org/10.1128/AEM.00246-06
Brodie, E. L., & Lynch, S. V. (2013). Phyloarrays. John Wiley & Sons, Ltd.
Castro-Ferreira, M. P., de Boer, T. E., Colbourne, J. K., Vooijs, R., van Gestel, C. A., van Straalen, N. M., Soares, A. M., Amorim, M. J., & Roelofs, D. (2014). Transcriptome assembly and microarray construction for Enchytraeus crypticus, a model oligochaete to assess stress response mechanisms derived from soil conditions. BMC Genomics, 15(1), 302. https://doi.org/10.1186/1471-2164-15-302
Cevher-Keskin, B., Yıldızhan, Y., Yüksel, B., Dalyan, E., & Memon, A. R. (2019). Characterization of differentially expressed genes to Cu stress in Brassica nigra by Arabidopsis genome arrays. Environmental Science and Pollution Research, 26(1), 299–311. https://doi.org/10.1007/s11356-018-3577-7
Chakraborty, R., Brodie, E. L., Nostrand, J. V., Zhou, J., & Hazen, T. C. (2008). Investigation of Cr (VI) tolerant bacteria from Cr (VI)-contaminated 100H site at Hanford, WA.
Chandler, D. P., Kukhtin, A., Mokhiber, R., Knickerbocker, C., Ogles, D., Rudy, G., Golova, J., Long, P., & Peacock, A. (2010). Monitoring microbial community structure and dynamics during in situ U (VI) bioremediation with a field-portable microarray analysis system. Environmental Science & Technology, 44(14), 5516–5522. https://doi.org/10.1021/es1006498
Cindy, S. (2015). Development of microarrays-based metagenomics technology for monitoring sulfate-reducing bacteria in subsurface environments. Glomics Inc.
Cong, J., Liu, X., Lu, H., Xu, H., Li, Y., Deng, Y., Li, D., & Zhang, Y. (2015). Analyses of the influencing factors of soil microbial functional gene diversity in tropical rainforest based on GeoChip 5.0. Genom Data, 5(5), 397–398. https://doi.org/10.1016/j.gdata.2015.07.010
Dalyan, E., Yüzbaşıoğlu, E., Keskin, B. C., Yıldızhan, Y., Memon, A., Ünal, M., & Yüksel, B. (2017). The identification of genes associated with Pb and Cd response mechanism in Brassica juncea L. by using Arabidopsis expression array. Environmental and Experimental Botany, 139, 105–115. https://doi.org/10.1016/j.envexpbot.2017.05.001
Dick, R., Muriel, D. B., Valeria, A., Pascal, B., Juliette, L., & Nico, V. S. (2012). Functional environmental genomics of a municipal landfill soil. Frontiers in Genetics. https://doi.org/10.3389/fgene.2012.00085
Epelde, L., Becerril, J. M., Kowalchuk, G. A., Deng, Y., Zhou, J., & Garbisu, C. (2010). Impact of metal pollution and Thlaspi caerulescens growth on soil microbial communities. Applied and Environment Microbiology, 76(23), 7843–7853. https://doi.org/10.1128/AEM.01045-10
Eyers, L., George, I., Schuler, L., Stenuit, B., Agathos, S. N., & El Fantroussi, S. (2004). Environmental genomics: Exploring the unmined richness of microbes to degrade xenobiotics. Applied Microbiology and Biotechnology, 66(2), 123–130. https://doi.org/10.1007/s00253-004-1703-6
Fountain, M. T., & Hopkin, S. P. (2005). Folsomia candida (Collembola): a “standard" soil arthropod. Annual Review of Entomology, 50(1), 201–222. https://doi.org/10.1146/annurev.ento.50.071803.130331
Gans, J., Wolinsky, M., & Dunbar, J. (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science, 309(5739), 1387–1390. https://doi.org/10.1126/science.1112665
Gorfer, M., Persak, H., Berger, H., Brynda, S., Bandian, D., & Strauss, J. (2009). Identification of heavy metal regulated genes from the root associated ascomycete Cadophora finlandica using a genomic microarray. Mycological Research, 113(12), 1377–1388. https://doi.org/10.1016/j.mycres.2009.09.005
Gu, Y., Joy, D. V. N., Liyou, W., Zhili, H., Yujia, Q., Fang-Jie, Z., Jizhong, Z., & Hauke, S. (2017). Bacterial community and arsenic functional genes diversity in arsenic contaminated soils from different geographic locations. PLoS One, 12(5), e0176696. https://doi.org/10.1371/journal.pone.0176696
Hamadeh, H. K., Bushel, P. R., Jayadev, S., Martin, K., DiSorbo, O., Sieber, S., Bennett, L., Tennant, R., Stoll, R., Barrett, J. C., Blanchard, K., Paules, R. S., & Afshari, C. A. (2002). Gene expression analysis reveals chemical-specific profiles. Toxicological Sciences, 67(2), 2. https://doi.org/10.1093/toxsci/67.2.219
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J., & Goodman, R. M. (1998). Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chemistry & Biology. https://doi.org/10.1016/s1074-5521(98)90108-9
He, Z., Deng, Y., Van Nostrand, J. D., Tu, Q., Xu, M., Hemme, C. L., Li, X., Wu, L., Gentry, T. J., Yin, Y., Liebich, J., Hazen, T. C., & Zhou, J. (2010). GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME Journal, 4(9), 1167–1179. https://doi.org/10.1038/ismej.2010.46
He, Z., Gentry, T. J., Schadt, C. W., Wu, L., Liebich, J., Chong, S. C., Huang, Z., Wu, W., Gu, B., Jardine, P., Criddle, C., & Zhou, J. (2007). GeoChip: A comprehensive microarray for investigating biogeochemical, ecological and environmental processes. The ISME Journal, 1(1), 67–77. https://doi.org/10.1038/ismej.2007.2
Hirano, T., & Tamae, K. (2011). Earthworms and soil pollutants. Sensors, 11(12), 11157–11167. https://doi.org/10.3390/s111211157
Hu, P., Brodie, E. L., Suzuki, Y., McAdams, H. H., & Andersen, G. L. (2005). Whole-genome transcriptional analysis of heavy metal stresses in caulobacter crescentus. Journal of Bacteriology, 187(24), 8437–8449. https://doi.org/10.1128/JB.187.24.8437-8449.2005
Jean-Marie, R., Michael, Z., & Erdogan, G. (2003). OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Research, 31(12), 3057–3062. https://doi.org/10.1093/nar/gkg426
Jiang, Z., Li, P., Wang, Y., Liu, H., Wei, D., Yuan, C., & Wang, H. (2019). Arsenic mobilization in a high arsenic groundwater revealed by metagenomic and Geochip analyses. Scientific Reports. https://doi.org/10.1038/s41598-019-49365-w
Jie, S., Li, M., Gan, M., Zhu, J., Yin, H., & Liu, X. (2016). Microbial functional genes enriched in the Xiangjiang River sediments with heavy metal contamination. BMC Microbiology, 16(1), 179. https://doi.org/10.1186/s12866-016-0800-x
Jin, M., & Li, J. W. (2008). Microarray application in environmental microbial community research. Microbiology China, 35(9), 1466–1471.
Kang, S., Van, N. J. D., Gough, H. L., He, Z., Hazen, T. C., Stahl, D. A., & Zhou, J. (2013). Functional gene array–based analysis of microbial communities in heavy metals-contaminated lake sediments. Fems Microbiology Ecology, 86(2), 2. https://doi.org/10.1111/1574-6941.12152
Kawata, K., Yokoo, H., Shimazaki, R., & Okabe, S. (2007). Classification of heavy-metal toxicity by human DNA microarray analysis. Environmental Science & Technology, 41(10), 3769–3774. https://doi.org/10.1021/es062717d
Kuramae, E. E., Yergeau, E., Wong, L. C., Pijl, A. S., van Veen, J. A., & Kowalchuk, G. A. (2012). Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiology Ecology, 79(1), 12–24. https://doi.org/10.1111/j.1574-6941.2011.01192.x
Lemos, L. N., Fulthorpe, R. R., Triplett, E. W., & Roesch, L. F. (2011). Rethinking microbial diversity analysis in the high throughput sequencing era. Journal of Microbiological Methods, 86(1), 42. https://doi.org/10.1016/j.mimet.2011.03.014
Leydesdorff, L., Carley, S., & Rafols, I. (2012). Global maps of science based on the new web-of-science categories. Scientometrics, 94(2), 589–593. https://doi.org/10.1007/s11192-012-0784-8
Liang, Y., He, Z., Wu, L., Deng, Y., Li, G., & Zhou, J. (2010). Development of a common oligonucleotide reference standard for microarray data normalization and comparison across different microbial communities. Applied & Environmental Microbiology, 76(4), 1088–1094. https://doi.org/10.1128/AEM.02749-09
Madigan, M. T., Martinko, J. M., & Parker, J. (1996). Brock Biology of Microorganisms.
Memon, A. R., Yildizhan, Y., & Keskin, B. (2008). Phytoremediation of heavy metals from contaminated areas of Turkey. 4th European Bioremediation Conference Sep. pp. 3–6.
Mergeay, M., Monchy, S., Vallaeys, T., Auquier, V., Benotmane, A., Bertin, P., Taghavi, S., Dunn, J., Van Der Lelie, D., & Wattiez, R. (2003). Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal S, Vallaeys. T. FEMS Microbiology Reviews, 27(2–3), 385–410. https://doi.org/10.1016/S0168-6445(03)00045-7
Migeon, A., Blaudez, D., Wilkins, O., Montanini, B., Campbell, M. M., Richaud, P., Thomine, S., & Chalot, M. (2010). Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cellular and Molecular Life Sciences, 67(22), 3763–3784. https://doi.org/10.1007/s00018-010-0445-0
Monsieurs, P., Moors, H., Van Houdt, R., Janssen, P. J., Janssen, A., Coninx, I., Mergeay, M., & Leys, N. (2011). Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. BioMetals, 24(6), 1133–1151. https://doi.org/10.1007/s10534-011-9473-y
Montero-Palmero, M. B. (2014). Environmental impact of toxic metals and metalloids. The case of mercury. In: A transcriptomic study of the early responses of Medicago sativa to mercury, pp. 3.
Moore, C. M., Gaballa, A., Hui, M., Ye, R. W., & Helmann, J. D. (2005). Genetic and physiological responses of Bacillus subtilis to metal ion stress. Molecular Microbiology, 57(1), 27–40. https://doi.org/10.1111/j.1365-2958.2005.04642.x
Nelson, D. L., & Cox, M. M. (2005). Lehninger Principles of Biochemistry. Lehninger principles of biochemistry.
Nota, B., Timmermans, M. J., Franken, O., Montagne-Wajer, K., Mariën, J., Boer, M. E. D., Boer, T. E. D., Ylstra, B., Straalen, N. M. V., & Roelofs, D. (2008). Gene expression analysis of collembola in cadmium containing soil. Environmental Science & Technology, 42(21), 8152–8157. https://doi.org/10.1021/es801472r
Nota, B., Verweij, R. A., Molenaar, D., Ylstra, B., van Straalen, N. M., & Roelofs, D. (2010). Gene expression analysis reveals a gene set discriminatory to different metals in soil. Toxicological Sciences, 115(1), 34–40. https://doi.org/10.1093/toxsci/kfq043
Ochsner, U. A., Wilderman, P. J., Vasil, A. I., & Vasil, M. L. (2002). GeneChip® expression analysis of the iron starvation response in Pseudomonas aeruginosa: Identification of novel pyoverdine biosynthesis genes. Molecular Microbiology, 45(5), 1277–1287. https://doi.org/10.1046/j.1365-2958.2002.03084.x
Olsson-Francis, K., Van Houdt, R., Mergeay, M., Leys, N., & Cockell, C. S. (2010). Microarray analysis of a microbe-mineral interaction. Geobiology, 8(5), 446–456. https://doi.org/10.1111/j.1472-4669.2010.00253.x
Pathak, A., Shanker, R., Garg, S. K., & Manickam, N. (2011). Profiling of biodegradation and bacterial 16S rRNA genes in diverse contaminated ecosystems using 60-mer oligonucleotide microarray. Applied Microbiology and Biotechnology, 90(5), 1739–1754. https://doi.org/10.1007/s00253-011-3268-5
Plessl, M., Rigola, D., Hassinen, V., Aarts, M. G., Schat, H., & Ernst, D. (2005). Transcription profiling of the metal-hyperaccumulator Thlaspi caerulescens (J. & C. PRESL). Zeitschrift Für Naturforschung C, 60(3–4), 216–223. https://doi.org/10.1515/znc-2005-3-406
Rastogi, G., Barua, S., Sani, R. K., & Peyton, B. M. (2011). Investigation of microbial populations in the extremely metal-contaminated Coeur d’Alene River sediments. Microbial Ecology, 62(1), 1–13. https://doi.org/10.1007/s00248-011-9810-2
Rastogi, G., Osman, S., Vaishampayan, P. A., Andersen, G. L., Stetler, L. D., & Sani, R. K. (2010). Microbial diversity in uranium mining-impacted soils as revealed by high-density 16S microarray and clone library. Microbial Ecology, 59(1), 94–108. https://doi.org/10.1007/s00248-009-9598-5
Rivas, L. A., García-Villadangos, M., Moreno-Paz, M., Cruz-Gil, P., Gómez-Elvira, J., & Parro, V. (2008). A 200-antibody microarray biochip for environmental monitoring: Searching for universal microbial biomarkers through immunoprofiling. Analytical Chemistry, 80(21), 7970–7979. https://doi.org/10.1021/ac8008093
Roelofs, D., Janssens, T. K., Timmermans, M. J., Nota, B., Marien, J., Bochdanovits, Z., Ylstra, B., & Van Straalen, N. M. (2009). Adaptive differences in gene expression associated with heavy metal tolerance in the soil arthropod Orchesella cincta. Molecular Ecology, 18(15), 3227–3239. https://doi.org/10.1111/j.1365-294X.2009.04261.x
Roh, S. W., Abell, G. C. J., Kim, K. -H., Nam, Y. -D., & Bae, J. -W. (2010). Comparing microarrays and next-generation sequencing technologies for microbial ecology research. Trends in Biotechnology, 28(6), 291–299. https://doi.org/10.1016/j.tibtech.2010.03.001
Roy, S. V., Vanbroekhoven, K., Dejonghe, W., & Diels, L. (2006). Immobilization of heavy metals in the saturated zone by sorption and in situ bioprecipitation processes. Hydrometallurgy, 83(1), 195–203. https://doi.org/10.1016/j.hydromet.2006.03.024
Sánchez-Fortún, M., Ouled-Cheikh, J., Jover, C., García-Tarrasón, M., Carrasco, J. L., & Sanpera, C. (2020). Following up mercury pollution in the Ebro Delta (NE Spain): Audouin’s gull fledglings as model organisms to elucidate anthropogenic impacts on the environment. Environmental Pollution, 266, 115232. https://doi.org/10.1016/j.envpol.2020.115232
Shalon, D., Smith, S. J., & Brown, P. O. (1996). A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Research, 6(7), 639–645. https://doi.org/10.1101/gr.6.7.639
Shen, L., Liu, X., & Qiu, G. (2008). Gene function and microbial community structure in sulfide minerals bioleaching system based on microarray analysis. Chinese Journal of Biotechnology, 24(6), 968–974. https://doi.org/10.3724/SP.J.1005.2008.00568
Singh, B. K., Quince, C., Macdonald, C. A., Khachane, A., Thomas, N., Al-Soud, W. A., Sørensen, S. J., He, Z., White, D., Sinclair, A., Crooks, B., Zhou, J., & Campbell, C. D. (2014). Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environmental Microbiology, 16(8), 2408–2420. https://doi.org/10.1111/1462-2920.12353
Sitte, J., Löffler, S., Burkhardt, E.-M., Goldfarb, K. C., Büchel, G., Hazen, T. C., & Küsel, K. (2015). Metals other than uranium affected microbial community composition in a historical uranium-mining site. Environmental Science and Pollution Research, 22(24), 19326–19341. https://doi.org/10.1007/s11356-015-4791-1
Small, J., Call, D. R., Brockman, F. J., Straub, T. M., & Chandler, D. P. (2001). Direct Detection of 16S rRNA in Soil Extracts by Using Oligonucleotide Microarrays. Applied & Environmental Microbiology, 67(10), 4708–4716. https://doi.org/10.1128/Aem.67.10.4708-4716.2001
Stone, J., Burgos, W., Royer, R., & Dempsey, B. (2006). Impact of zinc on biological Fe(III) and nitrate reduction by Shewanella putrefaciens CN32. Environmental Engineering Science, 23(4), 691–704. https://doi.org/10.1089/ees.2006.23.691
Sun, Y. J., & Zhang, H. C. (2013). Application of functional gene chip in research of soil microecology. South-to-North Water Transfers and Water Science & Technology, 11(01), 93–96.
Teng, Y., Luo, Y. M., & Li, Z. G. (2006). Microbial diversity in polluted soils: An overview. Acta Pedologica Sinica, 43(6), 1018–1026. https://doi.org/10.11766/trxb200512080620
Ueno, D., Milner, M. J., Yamaji, N., Yokosho, K., Koyama, E., Clemencia Zambrano, M., Kaskie, M., Ebbs, S., Kochian, L. V., & Ma, J. F. (2011). Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. The Plant Journal, 66(5), 852–862. https://doi.org/10.1111/j.1365-313X.2011.04548.x
Van Eck, N. J., & Waltman, L. (2010). Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics, 84(2), 523–538. https://doi.org/10.1007/s11192-009-0146-3
Wan, J., Tokunaga, T. K., Brodie, E., Wang, Z., Zheng, Z., Herman, D., Hazen, T. C., Firestone, M. K., & Sutton, S. R. (2005). Reoxidation of bioreduced uranium under reducing conditions. Environmental Science & Technology, 39(16), 6162–6169. https://doi.org/10.1021/es048236g
Wang, J. J., & Hou, P. Q. (2008). Gene chip technology. Preventive Medicine Tribune, 14(3), 285–287.
Weber, M., Harada, E., Vess, C., Roepenack-Lahaye, E. V., & Clemens, S. (2004). Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. The Plant Journal, 37(2), 269–281. https://doi.org/10.1046/j.1365-313X.2003.01960.x
Wei, Z., Hao, Z., Li, X., Guan, Z., Cai, Y., & Liao, X. (2019). The effects of phytoremediation on soil bacterial communities in an abandoned mine site of rare earth elements. Science of the Total Environment, 670, 950–960. https://doi.org/10.1016/j.scitotenv.2019.03.118
Wu, F., You, Y. Q., Werner, D., Jiao, S., Hu, J., Zhang, X. Y., Wan, Y., Liu, J. F., Wang, B., & Wang, X. L. (2020). Carbon nanomaterials affect carbon cycle-related functions of the soil microbial community and the coupling of nutrient cycles. Journal of Hazardous Materials. https://doi.org/10.1016/j.jhazmat.2020.122144
Xie, J., He, Z., Liu, X., Liu, X., Van Nostrand, J. D., Deng, Y., Wu, L., Zhou, J., & Qiu, G. (2011). GeoChip-based analysis of the functional gene diversity and metabolic potential of microbial communities in acid mine drainage. Applied & Environmental Microbiology, 77(3), 991–999. https://doi.org/10.1128/AEM.01798-10
Xiong, J., He, Z., Van Nostrand, J. D., Luo, G., Tu, S., Zhou, J., & Wang, G. (2012). Assessing the microbial community and functional genes in a vertical soil profile with long-term arsenic contamination. PLoS One, 7(11), e50507. https://doi.org/10.1371/journal.pone.0050507
Yokoi, T., Kaku, Y., Suzuki, H., Ohta, M., Ikuta, H., Isaka, K., Sumino, T., & Wagatsuma, M. (2007). “FloraArray” for screening of specific DNA probes representing the characteristics of a certain microbial community. FEMS Microbiology Letters, 273(2), 166–171. https://doi.org/10.1111/j.1574-6968.2007.00799.x
Yu, Z., He, Z., Tao, X., Zhou, J., Yang, Y., Zhao, M., Zhang, X., Zheng, Z., Yuan, T., & Liu, P. (2016). The shifts of sediment microbial community phylogenetic and functional structures during chromium (VI) reduction. Ecotoxicology, 25(10), 1759–1770. https://doi.org/10.1007/s10646-016-1719-6
Zhang, H. B., Duan, C. Q., & Qu, L. H. (2003). Culture independent methods for studies on microbial ecology of soils. Chinese Journal of Ecology, 22(5), 131–136.
Zhang, Y., Zhang, S. G., Qi, L. W., Chen, X. Q., Chen, R. Y., & Song, W. Q. (2006). Poplar as a model for Forest Tree in genome research. Chinese Bulletin of Botany.
Zhang, Y. G., Li, D. Q., & Xiao, Q. M. (2004). Microarrays and their application to environmental microorganisms. Acta Microbiologica Sinica, 44(3), 406–410. https://doi.org/10.1360/biodiv.050058
Zhou, J., He, Z., Yang, Y., Deng, Y., Tringe, S. G., & Alvarez-Cohen, L. (2015). High-throughput metagenomic technologies for complex microbial community analysis: Open and closed formats. Mbio, 6(1), e02288-e2314. https://doi.org/10.1128/mbio.02288-14
Zhou, J. Z., Xue, K., Xie, J. P., Deng, Y., & Luo, Y. (2012). Microbial mediation of carbon-cycle feedbacks to climate warming. Nature Climate Change, 2(2), 106–110. https://doi.org/10.1038/Nclimate1331
Zou, Y., Ning, D. L., Huang, Y., Liang, Y. T., Wang, H., Duan, L., Yuan, T., He, Z. L., Yang, Y. F., Xue, K., Van Nostrand, J. D., & Zhou, J. Z. (2020). Functional structures of soil microbial community relate to contrasting N2O emission patterns from a highly acidified forest. Science of the Total Environment. https://doi.org/10.1016/j.scitotenv.2020.138504
Funding
This work was supported by Key laboratory of Degraded and Unused Land Consolidation Engineering, and the Ministry of Natural and Resources (Grant No. SXDJ2019).
Author information
Authors and Affiliations
Contributions
HFS participated in the conception, design, data collection and analysis, and drafted the manuscript. ZCL assisted the laboratory work, results interpretation, and manuscript revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, Z., Su, H. Employing gene chip technology for monitoring and assessing soil heavy metal pollution. Environ Monit Assess 194, 2 (2022). https://doi.org/10.1007/s10661-021-09650-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-021-09650-6