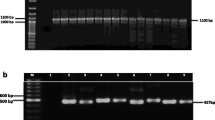
Abstract
Aquatic environments are hotspots for the spread of antibiotic-resistant bacteria and genes due to pollution caused mainly by anthropogenic activities. The aim of this study was to evaluate the impact of wastewater effluents, informal settlements, hospital, and veterinary clinic discharges on the occurrence, antibiotic resistance profile and virulence signatures of Aeromonas spp. and Pseudomonas spp. isolated from surface water and wastewater. High counts of Aeromonas spp. (2.5 (± 0.8) – 3.3 (± 0.4) log10 CFU mL−1) and Pseudomonas spp. (0.6 (± 1.0) – 1.8 (± 1.0) log10 CFU mL−1) were obtained. Polymerase chain reaction (PCR) and MALDI-TOF characterization identified four species of Aeromonas and five of Pseudomonas. The isolates displayed resistance to 3 or more antibiotics (71% of Aeromonas and 94% of Pseudomonas). Aeromonas spp. showed significant association with the antibiotic meropenem (χ2 = 3.993, P < 0.05). The virulence gene aer in Aeromonas was found to be positively associated with the antibiotic resistance gene blaOXA (χ2 = 6.657, P < 0.05) and the antibiotic ceftazidime (χ2 = 7.537, P < 0.05). Aeromonas recovered from both wastewater and surface water displayed high resistance to ampicillin and had higher multiple antibiotic resistance (MAR) indices close to the hospital. Pseudomonas isolates on the other hand exhibited low resistance to carbapenems but very high resistance to the third-generation cephalosporins and cefixime. The results showed that some of the Pseudomonas spp. and Aeromonas spp. isolates were extended-spectrum β-lactamase producing bacteria. In conclusion, the strong association between virulence genes and antibiotic resistance in the isolates shows the potential health risk to communities through direct and indirect exposure to the water.


Similar content being viewed by others
References
Abebe, E., Tegegne, B., Tibebu, S. J. E., et al. (2016). A review on molecular mechanisms of bacterial resistance to antibiotics. European Journal of Applied Sciences, 8(5), 301–310.
Abdelraouf, K., Kabbara, S., Ledesma, K. R., Poole, K., Tam, V. H., et al. (2011). Effect of multidrug resistance-conferring mutations on the fitness and virulence of Pseudomonas aeruginosa. Journal of Antimicrobial Chemotherapy, 66(6), 1311–1317.
Adegoke, A. A., Faleye, A. C., Singh, G., Stenström, T. A., et al. (2016). Antibiotic resistant superbugs: Assessment of the interrelationship of occurrence in clinical settings and environmental niches. Molecules. https://doi.org/10.3390/molecules22010029
Adeosun, F., Adams, T., Amrevuawho, M., et al. (2016). Effect of anthropogenic activities on the water quality parameters of Federal University of Agriculture Abeokuta reservoir. International Journal of Fisheries and Aquatic Sciences, 4(3), 104–108.
Agersø, Y. & Sandvang, D. (2005). Class 1 integrons and tetracycline resistance genes in Alcaligenes, Arthrobacter, and Pseudomonas spp. isolated from pigsties and manured soil. Applied and Environmental Microbiology, 71(12), 7941–7947.
Ajayi, T., Allmond, L. R., Sawa, T., Wiener-Kronish, J. P., et al. (2003). Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. Journal of Clinical Microbiology, 41(8), 3526–3531.
Allydice-Francis, K. & Brown, P. D. (2012). Diversity of antimicrobial resistance and virulence determinants in Pseudomonas aeruginosa associated with fresh vegetables. International Journal of Microbiology. https://doi.org/10.1155/2012/426241
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J., et al. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410.
Anderson, N. W., Buchan, B. W., Riebe, K. M., Parsons, L. N., Gnacinski, S., Ledeboer, N. A., et al. (2012). Effects of solid-medium type on routine identification of bacterial isolates by use of matrix-assisted laser desorption ionization time of flight mass spectrometry. Journal of Clinical Microbiology, 50(3), 1008–1013.
Aybey, A., & Demirkan, E. (2016). Inhibition of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa by human serum paraoxonase. Journal of Medical Microbiology, 65(2), 105–113.
Barrios-Hernández, M. L., Pronk, M., Garcia, H., Boersma, A., Brdjanovic, D., van Loosdrecht, M. C., & Hooijmans, M. C. (2020). Removal of bacterial and viral indicator organisms in full-scale aerobic granular sludge and conventional activated sludge systems. Water Research X, 6, 100040.
Batrich, M., Maskeri, L., Schubert, R., Ho, B., Kohout, M., Abdeljaber, M., Abuhasna, A., Kholoki, M., Psihogios, P., Razzaq, T., et al. (2019). Pseudomonas diversity within urban freshwaters. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2019.00195
Bergmark, L., Poulsen, P. H. B., Al-Soud, W. A., Norman, A., Hansen, L. H., Sørensen, S. J., et al. (2012). Assessment of the specificity of Burkholderia and Pseudomonas qPCR assays for detection of these genera in soil using 454 pyrosequencing. FEMS Microbiology Letters, 333(1), 77–84.
Bhatia, N., Castro-Borobio, M., Greene, J. N., Nanjappa, S., et al. (2017). Necrotizing fasciitis secondary to Aeromonas infection presenting with septic shock. Case Reports in Medicine. https://doi.org/10.1155/2017/4607582
Bitsori, M., Maraki, S., Koukouraki, S., Galanakis, E., et al. (2012). Pseudomonas aeruginosa urinary tract infection in children: Risk factors and outcomes. The Journal of Urology, 187(1), 260–264.
Bjarnsholt, T., Jensen, P. Ø., Fiandaca, M. J., Pedersen, J., Hansen, C. R., Andersen, C. B., Pressler, T., Givskov, M., Høiby, N., et al. (2009). Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatric Pulmonology, 44(6), 547–558.
Blasco, M., Esteve, C., Alcaide, E., et al. (2008). Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. Journal of Applied Microbiology, 105(2), 469–475.
Bradbury, R. S., Roddam, L., Merritt, A., Reid, D. W., Champion, A. C., et al. (2010). Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. Journal of Medical Microbiology, 59(8), 881–890.
Bratu, S., Gupta, J., Quale, J., et al. (2006). Expression of the las and rhl quorum-sensing systems in clinical isolates of Pseudomonas aeruginosa does not correlate with efflux pump expression or antimicrobial resistance. Journal of Antimicrobial Chemotherapy, 58(6), 1250–1253.
Braune, E., & Xu, Y. (2008). Groundwater management issues in Southern Africa: An IWRM perspective. Water SA, 34(6), 699–706.
Codjoe, F., Donkor, E. S., et al. (2018). Carbapenem resistance: A review. Journal of Medical Sciences. https://doi.org/10.3390/medsci6010001
CLSI. (2017). Performance standards for antimicrobial susceptibility testing; 27th information supplement M100–S27. . Clinical and Laboratory Standards Institute.
CLSI. (2016). Methods for antimicrobial dilutions and disk susceptibility testing; 3rd information supplement M45. Clinical and Laboratory Standards Institute.
Cherkaoui, A., Emonet, S., Fernandez, J., Schorderet, D., Schrenzel, J., et al. (2011). Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of beta-hemolytic Streptococci. Journal of Clinical Microbiology, 49(8), 3004–3005.
Cirauqui, N., Abriata, L. A., Van der Goot, F. G., Dal Peraro, M. J. S. R., et al. (2017). Structural, physicochemical and dynamic features conserved within the aerolysin pore-forming toxin family. Scientific Reports. https://doi.org/10.1038/s41598-017-13714-4
Croxatto, A., Prod’hom, G., Greub, G., et al. (2012). Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiology Reviews, 36(2), 380–407.
Daskalov, H. (2006). The importance of Aeromonas hydrophila in food safety. Food Control, 17(6), 474–483.
Dashti, A. A., Jadaon, M. M., Abdulsamad, A. M., Dashti, H. M., et al. (2009). Heat treatment of bacteria: A simple method of DNA extraction for molecular techniques. Kuwait Medical Journal, 41(2), 117–122.
Deng, J., Fu, L., Wang, R., Yu, N., Ding, X., Jiang, L., Fang, Y., Jiang, C., Lin, L., Wang, Y. J., et al. (2014). Comparison of MALDI-TOF MS, gene sequencing and the Vitek 2 for identification of seventy-three clinical isolates of enteropathogens. Journal of Thoracic Disease., 6(5), 539–544.
Delmani, F.A., Jaran, A. S., Al Tarazi, Y., Masaadeh, H., Zaki, O., et al. (2017). Characterization of ampicillin resistant gene (blaTEM-1) isolated from E. coli in Northern Jordan. Asian Journal Biomedical Pharmaceutical Science, 7(61), 11–15.
Ekwanzala, M. D., Dewar, J. B., Kamika, I., Momba, M. N. B., et al. (2018). Systematic review in South Africa reveals antibiotic resistance genes shared between clinical and environmental settings. Infection and Drug Resistance, 11, 1907–1920.
El Solh, A. A., & Alhajhusain, A. (2009). Update on the treatment of Pseudomonas aeruginosa pneumonia. Journal of Antimicrobial Chemotherapy, 64(2), 229–238.
Evangelista-Barreto, N. S., Carvalho, F. C. T. D., Vieira, R. H. S., Dos Reis, C. M. F., Macrae, A., Rodrigues, D. D. P., et al. (2010). Characterization of Aeromonas species isolated from an estuarine environment. Brazilian Journal of Microbiology, 41(2), 452–460.
Faraji, F., Mahzounieh, M., Ebrahimi, A., Fallah, F., Teymournejad, O., Lajevardi, B., et al. (2016). Molecular detection of virulence genes in Pseudomonas aeruginosa isolated from children with cystic fibrosis and burn wounds in Iran. Microbial Pathogenesis, 99, 1–4.
Fariñas, M. C., & Martínez-Martínez, L. (2013). Multiresistant Gram negative bacterial infections: Enterobacteria, Pseudomonas aeruginosa, Acinetobacter baumannii and other non-fermenting Gram negative bacilli. Enfermedades Infecciosas y Microbiologia Clinica, 31(6), 402–409.
Fazeli, N. & Momtaz, H. (2014). Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iranian Red Crescent Medical Journal. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4329751
Figueira, V., Vaz-Moreira, I., Silva, M., Manaia, C. M., et al. (2011). Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Research, 45(17), 5599–5611.
Fisher, J. C., Newton, R. J., Dila, D. K., McLellan, S. L., et al. (2015). Urban microbial ecology of a freshwater estuary of Lake Michigan. Elementa (Washington, DC). https://doi.org/10.12952/journal.elementa.000064
Garau, J., & Gomez, L. (2003). Pseudomonas aeruginosa pneumonia. Current Opinion in Infectious Diseases, 16(2), 135–143.
Garbis, H., Van Tonningen, M. R. & Reuvers, M. (2007). Anti-infective agents. Drugs During Pregnancy and Lactation (Second Edition). Elsevier, 123–177.
Gavin, R., Rabaan, A. A., Merino, S., Tomás, J. M., Gryllos, I., Shaw, J. G., et al. (2002). Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Molecular Microbiology, 43(2), 383–397.
Geisinger, E., & Isberg, R. R. (2017). Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. The Journal of Infectious Diseases, 215(1), S9–S17.
Golle, A., Janezic, S., & Rupnik, M. J. (2017). Low overlap between carbapenem resistant Pseudomonas aeruginosa genotypes isolated from hospitalized patients and wastewater treatment plants. PLoS One. https://doi.org/10.1371/journal.pone.0186736
Goñi-Urriza, M., Pineau, L., Capdepuy, M., Roques, C., Caumette, P., Quentin, C., et al. (2000). Antimicrobial resistance of mesophilic Aeromonas spp. isolated from two european rivers. Journal of Antimicrobial Chemotherapy, 46(2), 297–301.
Guo, J., Li, J., Chen, H., Bond, P. L., Yuan, Z., et al. (2017). Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Research, 123, 468–478.
Hassard, F., Gwyther, C. L., Farkas, K., Andrews, A., Jones, V., Cox, B., Brett, H., Jones, D. L., McDonald, J. E., Malham, S. K., et al. (2016). Abundance and distribution of enteric bacteria and viruses in coastal and estuarine sediments—A review. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2016.01692
Hembach, N., Schmid, F., Alexander, J., Hiller, C., Rogall, E. T., Schwartz, T., et al. (2017). Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2017.01282
Heuer, H., & Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environmental Microbiology, 9(3), 657–666.
Igbinosa, I. H., & Okoh, A. I. (2013). Antibiotic susceptibility profile of Aeromonas species isolated from wastewater treatment plant. The Scientific World Journal. https://doi.org/10.1100/2012/625023
Igbinosa, I. H., Nwodo, U. U., Sosa, A., Tom, M., Okoh, A. I., et al. (2012a). Commensal Pseudomonas species isolated from wastewater and freshwater milieus in the Eastern Cape Province, South Africa, as reservoir of antibiotic resistant determinants. International Journal of Environmental Research and Public Health, 9(7), 2537–2549.
Igbinosa, I. H., Igumbor, E. U., Aghdasi, F., Tom, M., Okoh, A. I., et al. (2012b). Emerging Aeromonas species infections and their significance in public health. The Scientific World Journal. https://doi.org/10.1100/2012/625023
Igbinosa, I. H., Igbinosa, E. O., Okoh, A. I., et al. (2014). Molecular detection of metallo-β-lactamase and putative virulence genes in environmental isolates of Pseudomonas species. Polish Journal of Environmental Studies, 23(6), 2327–2331.
Janda, J. M., & Abbott, S. L. (1998). Evolving concepts regarding the genus Aeromonas: An expanding panorama of species, disease presentations, and unanswered questions. Clinical Infectious Diseases, 27(2), 332–344.
Janda, J. M., & Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clinical Microbiology Reviews, 23(1), 35–73.
Jenkins, C. E., SwiatoniowskI, A., Issekutz, A. C., Lin, T. J., et al. (2004). Pseudomonas aeruginosa exotoxin A induces human mast cell apoptosis by a caspase 8 and 3 dependent mechanism. Journal of Biological Chemistry, 279(35), 37201–37207.
Juan, N. C., & Oliver, A. (2010). Carbapenemases in Pseudomonas spp. Enfermedades Infecciosas y Microbiologia Clinica, 28, 19–28.
Kaur, P., & Peterson, E. J. (2018). Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2018.02928
Kerrn, M., Klemmensen, T., Frimodt-Møller, N., Espersen, F., et al. (2002). Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. Journal of Antimicrobial Chemotherapy, 50(4), 513–516.
Kim, S. E., Park, S. H., Park, H. B., Park, K. H., Kim, S. H., Jung, S. I., Shin, J. H., Jang, H. C., Kang, S. J., et al. (2012). Nosocomial Pseudomonas putida bacteremia: High rates of carbapenem resistance and mortality. Chonnam Medical Journal, 48(2), 91–95.
Kingombe, C. I. B., Huys, G., Tonolla, M., Albert, M. J., Swings, J., Peduzzi, R., Jemmi, T. J., et al. (1999). PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Applied Environmental Microbiology, 65(12), 5293–5302.
Kirov, S. M., Castrisios, M., Shaw, J. G., et al. (2004). Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infection and Immunity, 72(4), 1939–1945.
Kuo, H.Y., Yang, C.M., Lin, M.F., Cheng, W.L., Tien, N., Liou, M.L., et al. (2010). Distribution of blaOXA carrying imipenem-resistant Acinetobacter spp. in 3 hospitals in Taiwan. Diagnostic Microbiology and Infectious Disease, 66(2), 195–199.
Küpfer, M., Kuhnert, P., Korczak, B. M., Peduzzi, R., Demarta, A., et al. (2006). Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. International Journal of Systematic and Evolutionary microbiology, 56(12), 2743–2751.
Lavenir, R., Jocktane, D., Laurent, F., Nazaret, S., Cournoyer, B., et al. (2007). Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target. Journal of Microbiological Methods, 70(1), 20–29.
Lee, S., Suits, M., Witusynski, D., Winston, R., Martin, J., Lee, J., et al. (2020). Residential urban stormwater runoff: A comprehensive profile of microbiome and antibiotic resistance. Science of The Total Environment. https://doi.org/10.1016/j.scitotenv.2020.138033
Li, D., Yang, M., Hu, J., Zhang, J., Liu, R., Gu, X., Zhang, Y., Wang, Z., et al. (2009). Antibiotic resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environmental Microbiology, 11(6), 1506–1517.
Li, X. Z., PlésiaT, P., Nikaido, H., et al. (2015). The challenge of efflux-mediated antibiotic resistance in Gram negative bacteria. Clinical Microbiology Reviews, 28(2), 337–418.
Livermore, D. M. (2002). Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our worst nightmare? Clinical Infectious Diseases, 34(5), 634–640.
Livermore, D. M., & Woodford, N. (2006). The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends in Microbiology, 14(9), 413–420.
Mackintosh, G., & Colvin, C. (2003). Failure of rural schemes in South Africa to provide potable water. Environmental Geology, 44(1), 101–105.
Marchesi, J. R., Sato, T., Weightman, A. J., Martin, T. A., Fry, J. C., Hiom, S. J., Wade, W. G., et al. (1998). Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Applied Environmental Microbiology, 64(2), 795–799.
Martínez, O., Rodríguez-Calleja, J. M., Santos, J. A., Otero, A., García-López, M. L., et al. (2009). Foodborne and indicator bacteria in farmed molluscan shellfish before and after depuration. Journal of Food Protection, 72(7), 1443–1449.
Martone-Rocha, S., Piveli, R., Matté, G., Dória, M., Dropa, M., Morita, M., Peternella, F., Matté, M., et al. (2010). Dynamics of Aeromonas species isolated from wastewater treatment system. Journal of Water and Health, 8(4), 703–711.
Mema, V. (2010). Impact of poorly maintained waste water and sewage treatment plants: Lessons from South Africa. Resource, 12, 60–65.
Mena, K. D., & Gerba, C. P. (2009). Risk assessment of Pseudomonas aeruginosa in water. Reviews of Environmental Contamination and Toxicology, 201, 71–115.
Mendelson, M., & Matsoso, M. J. A. (2015). The South African antimicrobial resistance strategy framework. South African Medical Journal. https://doi.org/10.7196/SAMJ.9644
Michalska, M. & Wolf, P. (2015). Pseudomonas Exotoxin A: Optimized by evolution for effective killing. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2015.00963
Mudau, M., Jacobson, R., Minenza, N., Kuonza, L., Morris, V., Engelbrecht, H., Nicol, M. P., Bamford, C. J., et al. (2013). Outbreak of multi-drug resistant Pseudomonas aeruginosa bloodstream infection in the haematology unit of a South African academic hospital. PLoS One. https://doi.org/10.1371/journal.pone.0055985
Mulamattathil, S. G., Bezuidenhout, C., Mbewe, M., Ateba, C. N., et al. (2014). Isolation of environmental bacteria from surface and drinking water in Mafikeng, South Africa, and characterization using their antibiotic resistance profiles. Journal of Pathogens. https://doi.org/10.1155/2014/371208
Naidoo, S. & Olaniran, A. (2014). Treated wastewater effluent as a source of microbial pollution of surface water resources. Environmental Research and Public Health. https://doi.org/10.3390/ijerph110100249
Narciso-da-Rocha, C., Varela, A. R., Schwartz, T., Nunes, O. C., Manaia, C. M., et al. (2014). blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital–urban wastewater treatment plant system. Journal of Global Antimicrobial Resistance, 2(4), 309–315.
Nawaz, M., Khan, S. A., Khan, A. A., Sung, K., Tran, Q., Kerdahi, K., Steele, R.J., et al. (2010). Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiology. https://doi.org/10.1016/j.fm.2009.11.00
Nkuna, Z., Mamakoa, E., Mothetha, M., et al. (2014). The important role of springs in South Africa’s rural water supply: The case study of two rural communities in South Africa. OIDA International Journal of Sustainable Development, 7(12), 11–20.
Numberger, D., Ganzert, L., Zoccarato, L., Mühldorfer, K., Sauer, S., Grossart, H. P., Greenwood, A. D., et al. (2019). Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Scientific Reports. https://doi.org/10.1038/s41598-019-46015-z
Odeyemi, O. A. & Ahmad, A. J. (2017). Antibiotic resistance profiling and phenotyping of Aeromonas species isolated from aquatic sources. Saudi Journal of Biological Sciences. https://doi.org/10.1016/j.fm.2009.11.00
Odjadjare, E. E., Igbinosa, E. O., Mordi, R., Igere, B., Igeleke, C. L., Okoh, A. I., et al. (2012). Prevalence of multiple antibiotics resistant (MAR) Pseudomonas species in the final effluents of three municipal wastewater treatment facilities in South Africa. International Journal of Environmental Research and Public Health, 9(6), 2092–2107.
Olaniran, A. O., Nzimande, S. B., Mkize, N. G., et al. (2015). Antimicrobial resistance and virulence signatures of Listeria and Aeromonas species recovered from treated wastewater effluent and receiving surface water in Durban, South Africa. BMC Microbiology. https://doi.org/10.1186/s12866-015-0570-x
Olds, H. T., Corsi, S. R., Dila, D. K., Halmo, K. M., Bootsma, M. J., McLellan, S. L., et al. (2018). High levels of sewage contamination released from urban areas after storm events: A quantitative survey with sewage specific bacterial indicators. PLoS Medicine. https://doi.org/10.1371/journal.pmed.1002614
Oliver, D. M., Clegg, C. D., Heathwaite, A. L., Haygarth, P. M., et al. (2007). Preferential attachment of Escherichia coli to different particle size fractions of an agricultural grassland soil. Water, Air, and Soil Pollution. https://doi.org/10.1007/s11270-007-9451-8
Osundiya, O., Oladele, R., Oduyebo, O., et al. (2013). Multiple antibiotic resistance (MAR) indices of Pseudomonas and Klebsiella species isolates in Lagos University Teaching Hospital. African Journal of Clinical and Experimental Microbiology, 14(3), 164–168.
Pang, Z., Raudonis, R., Glick, B. R., Lin, T. J., Cheng, Z., et al. (2019). Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnology Advances, 37(1), 177–192.
Paterson, D. L., & Harris, P. N. A. (2016). Colistin resistance: A major breach in our last line of defence. Lancet Infectious Diseases, 16(2), 132–133.
Paul, P., Adikesavalu, H., Banerjee, S., Abraham, T. J., et al. (2015). Antibiotic resistant motile Aeromonads induced septicemia in Philippine catfish Clarias batrachus (Linnaeus, 1758) fingerlings. Croatian Journal of Fisheries, 73(4), 170–175.
Paul, S., Bezbaruah, R., Roy, M., Ghosh, A., et al. (1997). Multiple antibiotic resistance (MAR) index and its reversion in Pseudomonas aeruginosa. Letters in Applied Microbiology, 24(3), 169–171.
Pérez‐Valdespino, A., Fernández-Rendón, E., Curiel-Quesada, E., et al. (2009). Detection and characterization of class 1 integrons in Aeromonas spp. isolated from human diarrheic stool in Mexico. Journal of Basic Microbiology, 49(6), 572–578.
Pfeifer, Y., Cullik, A., Witte, W., et al. (2010). Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. International Journal of Medical Microbiology, 300(6), 371–379.
Piotrowska, M. & Popowska, M. (2015). Insight into the mobilome of Aeromonas strains. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2015.00494
Piotrowska, M., Kowalska, S., Popowska, M., et al. (2019). Diversity of β-lactam resistance genes in Gram-negative rods isolated from a municipal wastewater treatment plant. Annals of Microbiology, 69(6), 591-601.
Podobnik, M., Kisovec, M., Anderluh, G., et al. (2017). Molecular mechanism of pore formation by aerolysin-like proteins. Philosophical Transactions Royal Society. https://doi.org/10.1098/rstb.2016.0209
Prinsloo, A., van Straten, A. M. S., Weldhagen, G. F., et al. (2008). Antibiotic synergy profiles of multidrug-resistant Pseudomonas aeruginosa in a nosocomial environment. South African Journal of Epidemiology and Infection, 23(3), 7–9.
Ramírez-Castillo, F. Y., Loera-Muro, A., Jacques, M., Garneau, P., Avelar-González, F. J., Harel, J., Guerrero-Barrera, A. L., et al. (2015). Waterborne pathogens: Detection methods and challenges. Pathogens, 4(2), 307–334.
Rawat, D., & Nair, D. (2010). Extended-spectrum β-lactamases in Gram negative bacteria. Journal of Global Infectious Diseases, 2(3), 263–274.
Riaz, S., Faisal, M., Hasnain, S. J., et al. (2011). Antibiotic susceptibility pattern and multiple antibiotic resistances (MAR) calculation of extended spectrum β-lactamase (ESBL) producing Escherichia coli and Klebsiella species in Pakistan. African Journal of Biotechnology, 10(33), 6325–6331.
Robertson, B. K., Harden, C., Selvaraju, S. B., Pradhan, S., Yadav, J. S., et al. (2014). Molecular detection, quantification, and toxigenicity profiling of Aeromonas spp. in source and drinking water. The Open Microbiology Journal, 8, 32–39.
Ruiz, L. D., Domínguez, M. A., Ruiz, N., Viñas, M., et al. (2004). Relationship between clinical and environmental isolates of Pseudomonas aeruginosa in a hospital setting. Archives of Medical Research, 35(3), 251–257.
Ruiz-Villalba, A., van Pelt-Verkuil, E., Gunst, Q. D., Ruijter, J. M., van den Hoff, M. J., et al. (2017). Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR). Biomolecular Detection and Quantification, 14, 7–18.
Saka, B., Adeyemo, O., Odeseye, A., et al. (2017). Multiple antibiotic resistance indices of Aeromonas hydrophila isolates of muscle of catfish (Clarias gariepinus, Burchell 1822) from selected markets in Ibadan, Nigeria. African Journal of Clinical and Experimental Microbiology, 18(2), 73–78.
Sánchez, P., Linares, J. F., Ruiz-Díez, B., Campanario, E., Navas, A., Baquero, F., Martínez, J. L., et al. (2002). Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. Journal of Antimicrobial Chemotherapy, 50(5), 657–664.
Scales, B. S., Dickson, R. P., Lipuma, J. J., Huffnagle, G. B., et al. (2014). Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clinical Microbiology Reviews, 27(4), 927–948.
Sen, K., & Rodgers, M. (2004). Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: A PCR identification. Journal of Applied Microbiology, 97(5), 1077–1086.
Stats, S. A. (2017). Mid year population estimates (Statistical release P0302). Statistics South Africa.
Tam, V. H., Rogers, C. A., Chang, K. T., Weston, J. S., Caeiro, J. P., Garey, K. W., et al. (2010). Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrobial Agents and Chemotherapy, 54(9), 3717–3722.
Tao, R., Ying, G. G., Su, H. C., Zhou, H. W., Sidhu, J. P., et al. (2010). Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl Rivers in South China. Environmental Pollution, 158(6), 2101–2109.
Thenmozhi, S., Moorthy, K., Sureshkumar, B., Suresh, M., et al. (2014). Antibiotic resistance mechanism of ESBL producing Enterobacteriaceae in clinical field: A review. International Journal of Pure Applied Bioscience, 2(3), 207–226.
Thomas, B. S., Okamoto, K., Bankowski, M. J., Seto, T. B., et al. (2013). A lethal case of Pseudomonas putida bacteremia due to soft tissue infection. Infectious Diseases in Clinical Practice, 21(3), 147–213.
Timperio, A. M., Gorrasi, S., Zolla, L., Fenice, M. J., et al. (2017). Evaluation of MALDI-TOF mass spectrometry and MALDI BioTyper in comparison to 16S rDNA sequencing for the identification of bacteria isolated from Arctic sea water. PLoS One. https://doi.org/10.1371/journal.pone.0181860
Tomás, J. (2012). The main Aeromonas pathogenic factors. International Scholarly Research Network Microbiology. https://doi.org/10.5402/2012/256261/
Wang, C., Cai, P., Chang, D., Mi, Z., et al. (2006). A Pseudomonas aeruginosa isolate producing the GES-5 extended-spectrum β-lactamase. Journal of Antimicrobial Chemotherapy, 57(6), 1261–1262.
Warburton, D. W., Bowen, B., Konkle, A., et al. (1994). The survival and recovery of Pseudomonas aeruginosa and its effect upon Salmonellae in water: methodology to test bottled water in Canada. Canadian Journal of Microbiology, 40(12), 987–992.
World Health Organisation, (WHO). (2016). Protecting surface water for health: Identifying, assessing and managing drinking-water quality risks in surface-water catchments. http://www.who.int/water_sanitation_health/publications/pswh/en/ [Accessed 24–06–2020].
World Health Organisation, (WHO). (2017). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO [Accessed 24–05–2019].
Yang, C. H., Lee, S., Su, P. W., Yang, C. S., Chuang, L. Y., et al. (2008). Genotype and antibiotic susceptibility patterns of drug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii isolates in Taiwan. Microbial Drug Resistance, 14(4), 281–288.
Yoshino, Y., Kitazawa, T., Kamimura, M., Tatsuno, K., Ota, Y., Yotsuyanagi, H., et al. (2011). Pseudomonas putida bacteremia in adult patients: Five case reports and a review of the literature. Journal of Infection and Chemotherapy, 17(2), 278–282.
Yousefi-Avarvand, A., Khashei, R., Ebrahim-Saraie, H. S., Emami, A., Zomorodian, K., Motamedifar, M., et al. (2015). The frequency of exotoxin A and exoenzymes S and U genes among clinical isolates of Pseudomonas aeruginosa in Shiraz, Iran. International Journal of Molecular and Cellular Medicine, 4(3), 167.
Zhang, L., Levy, K., Trueba, G., Cevallos, W., Trostle, J., Foxman, B., Marrs, C. F., Eisenberg, J. N., et al. (2015). Effects of selection pressure and genetic association on the relationship between antibiotic resistance and virulence in Escherichia coli. Antimicrobial Agents and Chemotherapy, 59(11), 6733–6740.
Zhiyong, Z., Xiaoju, L., Yanyu, G., et al. (2002). Aeromonas hydrophila infection: clinical aspects and therapeutic options. Reviews in Medical Microbiology, 13(4), 151–162.
Acknowledgements
We would like to thank Dr Carl-Fredrik Flach and Maja Genheden from the Centre for Antibiotic Resistance Research (CARe) at University of Gothenburg, Sweden, for their assistance in identification of isolates using MALDI-TOF.
Funding
We are grateful to the South African Research Chair (SARChI) initiative of the National Research Foundation of South Africa (NRF) and the Department of Science and Technology for the research funds. One of the authors (RG) also received a Ph.D. scholarship from the NRF (grant number 91944). The funding institutions were not involved in the study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Govender, R., Amoah, I.D., Adegoke, A.A. et al. Identification, antibiotic resistance, and virulence profiling of Aeromonas and Pseudomonas species from wastewater and surface water. Environ Monit Assess 193, 294 (2021). https://doi.org/10.1007/s10661-021-09046-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-021-09046-6