Abstract
A nematological survey was conducted between 2021 and 2022 in banana fields distributed in two major banana-producing regions in the western coast of Syria. As a result, six populations of Xiphinema spp. identified as X. diffusum, X. pachtaicum, X. simile, X. vuittenezi and X. zagrosense were isolated from the rhizosphere of banana plants (Musa sp.) and characterized using morphological and molecular data based on two rRNA genes (D2–D3 expansion segments of the 28S, and ITS fragments) and partial region of the cytochrome oxidase I gene (COI mtDNA) sequences. Moreover, the molecular identification of the endosymbionts of these populations was also performed using the complete 16S rDNA gene. The phylogenetic relationships of the recovered species of nematodes and respective endosymbionts were reconstructed. Candidatus Xiphinematobacter sp. (OR196969; OR196971) and Ca. Xiphinematobacter sp. (OR196970) were detected in X. diffusum and X. simile, respectively, and clustered together with other Ca. Xiphinematobacter sp.A and sp.I respectively. To our knowledge, this is the first report of X. diffusum, X. simile, X. vuittenezi and X. zagrosense parasitizing banana in Syria, extending the geographical distribution of these species within the Mediterranean Basin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dorylaimida Pearse, 1942 is one of the most abundant and diverse orders within the phylum Nematoda Potts, 1932 (Potts, 1932; Pearse, 1942; Ahmed & Holovachov, 2021). The genus Xiphinema Cobb, 1913, commonly known as dagger nematode, belongs to the family Longidoridae Thorne, 1935 and to the suborder Dorylaimina Pearse, 1936 (Ahmed & Holovachov, 2021; Gutiérrez-Gutiérrez et al., 2016; Pearse, 1936). According to morphological and biological features, members of Xiphinema are currently divided into two sub-groups: (i) Xiphinema non-americanum group, comprising around 237 species (172 species listed in original key by Loof & Luc, 1990, 11 species in the first and 31 species in the second supplement by Loof and Luc (1993) and Loof et al. (1996); till 2019, 60 species added to the genus or revalidated as listed by Mirzaie Fouladvand et al., 2019, and five species added to the genus since 2019 by Jahanshahi Afshar et al., 2019; Vazifeh et al., 2019; Archidona-Yuste et al., 2020a, b; Cai et al., 2020; Poureskandarian et al., 2023), mainly characterized by a wide morphological diversity: having a body J- to C-shaped after heat relaxation, a relatively long body (usually among 3.0 and 8.5 mm long) and odontostyle, female with high variation in the reproductive system, with or without any type of uterine differentiations (i.e. Z or pseudo-Z organ, crystal-like and/or spine-like structures in the tubular part of the uterus) and a wide diversity of tail shapes (Coomans et al., 2001; Loof & Luc, 1990; Taylor & Brown, 1997); and (ii) Xiphinema americanum group, accounted with 60 valid species (49 species listed by Lamberti et al., 2000, 11 extra species listed by Mobasseri et al., 2019a, b; three species added by Jahanshahi Afshar et al., 2021; Naghavi et al., 2022a; Gu et al., 2022), characterized by a spiral body after heat relaxation, a relatively small body (usually among 1.2 and 3.0 mm long), a female reproductive system with two equally developed genital branches, short uteri without uterine differentiations, endosymbiotic bacteria in intestinal cells and in the reproductive system in some species, and a conical tail with sharp to broadly rounded tip (Coomans et al., 2001; Lamberti et al., 2000; Palomares-Rius et al., 2016, 2021). Xiphinema species cause severe damage to plants by their direct feeding on root cells (Jones et al., 2013; Taylor & Brown, 1997). However, some of them retain/transmit economically important nepoviruses (genus Nepovirus, family Comoviridae) (Taylor & Brown, 1997; Trudgill et al., 1983). Furthermore, some of these Xiphinema species are included in the list of quarantine organisms of the European and Mediterranean Plant Protection Organization (EPPO) (OEPP/EPPO, 2022) which emphasizes the need for an accurate nematode identification at the species level.
An integrative taxonomic approach is the most efficient and accurate strategy to overcome practical issues with dagger nematodes (Gutiérrez-Gutiérrez et al., 2010, 2011b; 2013; Archidona-Yuste et al., 2016a, b; 2020b; Palomares-Rius et al., 2017; Cai et al., 2019, 2020; Mobasseri et al., 2019a, b). Untill recently, Xiphinema spp. populations were usually only identified at the species-level using polytomous diagnostic keys (Lamberti et al., 2000, 2004; Loof & Luc, 1990; Loof et al., 1996). Nowadays, sequencing RNA-based markers represents a powerful approach for species-level taxonomic identification within the genus Xiphinema and understanding of the evolutionary relationships within this genus (Gutiérrez-Gutiérrez et al., 2010, 2011b; 2013; Archidona-Yuste et al., 2016a, b; 2019; Cai et al., 2018, 2019; Mobasseri et al., 2019a, b; Naghavi et al., 2022a).
Members of the X. americanum group harbour bacterial endosymbionts belonging to ‘Candidatus Xiphinematobacter’ and ‘Candidatus Xiphinematincola pachtaicus’ species (Palomares-Rius et al., 2016, 2021; Brown, 2018; Mobasseri et al., 2019a, b). These endosymbionts are vertically transmitted from mother to offspring (Brown, 2018; Palomares-Rius et al., 2016, 2021). The 16S rRNA gene is useful for studying the taxonomy and genetic diversity of bacterial endosymbionts and resolving the nematode-endosymbiont bacteria coevolution (Lazarova et al., 2016a; Mobasseri et al., 2019a, b; Orlando et al., 2016; Palomares-Rius et al., 2016, 2021).
Previous studies have reported the occurrence and geographical distribution of dagger nematodes in Syria and to date a total of six Xiphinema species have been described, namely X. americanum sensu lato (s.l.) Cobb, 1913, X. diversicaudatum (Micoletzky, 1927) Thorne, 1939, X. elongatum Schuurmans Stekhoven & Teunissen, 1939, X. index Thorne & Allen, 1950, X. italiae Meyl, 1953, and X. pachtaicum (Tulaganov, 1938) Kirjanova, 1951 (Abdine et al., 2007; Al-Halabi & Al-Assas, 2021; Ali, 2023; Allouf, 2013; Greco et al., 1984, 1992; Kawas & Al-Assas, 2011; Khaleel et al., 2010; Lamberti, 1984; Lamberti et al., 1999a).
A precise and accurate diagnosis of dagger nematode species in Syria is a crucial requisite for the selection of effective control methods. However, updated information on the biological diversity including DNA data, geographical occurrence, and distribution of dagger nematodes in banana (Musa sp.) plantations and their associated endosymbiont bacteria is lacking. Thus, the objectives of this work were the following: (1) to characterize and describe Xiphinema populations obtained from nematode surveys from two major banana-growing regions of the West Coast of Syria using integrative taxonomy; (2) to update information on the diversity of Xiphinema spp. recorded from Syria; (3) to establish phylogenetic relationships of the identified Xiphinema species with available sequenced species; and (4) to characterise the diversity and phylogeny of Candidatus Xiphinematobacter associated with populations of the X. americanum-group obtained from present survey.
Material and methods
Soil sampling and morphological studies
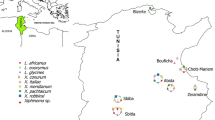
During two consecutive years (2021–2022) on the west coast of Syria, soil samples were collected from the rhizosphere of banana plants (Musa sp.) in 95 fields located in two of the main banana-growing regions (Latakia and Tartous). All sampling areas were arbitrarily chosen. Longidorid specimens were extracted from 500 cm3 soil samples using the modified Cobb washing, sieving and flotation technique (Flegg, 1967). As a result, 18 populations of Xiphinema were isolated from soil samples from which six populations were considered for further characterization (Table 1). The other populations showed low density (less than two juveniles were recovered).
After observation under a stereoscope, adult specimens of each morphotypes (or putative species) were hand-picked, and kept in both TAF fixative solution (Triethanolamine) and ethanol 70% (v/v) for morphological and molecular analyses, respectively. Subsequently, specimens maintained in TAF were processed to glycerol using Seinhorst’s method (Seinhorst, 1959) as modified by De Grisse (1969). The light micrographs and measurements of the nematode population, including the main diagnostic key characteristics [i.e., De Man indices (de Man, 1880), Loof-Coomans indices (Loof & Coomans, 1968), and other measurements and morphological features (Coomans et al., 2001; Lamberti et al., 2000, 2004; Loof & Luc, 1990)] were performed using an Olympus BX50 light microscope equipped with an Olympus DP70 digital camera (Olympus BX50, Hamburg, Germany) and Cell software (Olympus Software Imaging for Life Sciences). All measurements were expressed in micrometers (µm). All abbreviations are as defined in Jairajpuri and Ahmad (1992). Etymology used to describe the female genital branches is accodace with Coomans et al. (2001).
DNA sequencing and molecular methods
Total DNA isolation was performed using a thermodynamic and chemical approach according to Gutiérrez-Gutiérrez et al. (2020). DNA was extracted from a single individual adult specimen. Each individual nematode was placed in a 2 μL drop of sterile water on the flat cap of a PCR tube containing 20 µL of lysis buffer solution compounded by 10 µL ddH2O, 6 µL 10 × PCR buffer, 2 µL Mg2+ and 2 µL of proteinase K (20 mg/mL) (Nalgene®). Each specimen was cut into several very small pieces, usually between three and five pieces, with surface-sterilized and pointed pins. Next, tubes were centrifuged at 12,000 rpm for 30 s to 1 min and frozen at − 75 °C (for 60 min). Samples were mixed for 10 s, centrifuged at 12,000 rpm for 30 s to 1 min, and incubated at 57 °C (2 h), 65 °C (2 h) and 95 °C (15 min). PCR amplification was performed in a final volume of 25 μL containing: 1 µL of DNA template, 12.5 µL NZYTaq 2 × Green Master Mix (1.25 mM MgCl2, 200 mM dNTPs, 0.2 U/µL DNA Polymerase) (NZYTech, Lisbon, Portugal), 0.6 µL of each primer (10 mM), and 10.3 µL of ddH2O. Each rDNA (D2–D3 of 28S rRNA gene, the internal transcribed spacer 1 (ITS1) region, and 16S rRNA gene) and mtDNA (the partial portion of COI gene) fragment was amplified using one or several primer pairs (Table S1). All PCR assays were conducted as described by Gutiérrez-Gutiérrez et al. (2018, 2020). These PCR assays were conducted using DNA positive controls and a negative control containing sterile water. The PCR cycle conditions were followed as described by Gutiérrez-Gutiérrez et al. (2018, 2020) and Palomares-Rius et al. (2016, 2021). For a complete amplification of the 16S rRNA gene from the bacterial endosymbionts, specific PCR cycle conditions were used: initial denaturation at 95 °C for 3 min; followed by 30 amplification cycles of 94 °C for 30 s; an annealing temperature of 50 °C (27F/1492R; 27F/920R; 27F/800R) or 55 °C (360F/920R; 360F/1492R) for 30 s, 72 °C for 30–60 s; and final extension of 72 °C for 10 min. Each PCR product was purified and sequenced by STABVIDA (Caparica, Portugal). Additionally, 360R, 800R, 920R and 1100R primers (Table S1) were used to guarantee a robust and complete sequence. All new sequences of the Syrian populations of dagger nematode species were deposited into the GenBank database under the accession numbers indicated on the Table 1.
Molecular phylogenetic analyses
The newly obtained rDNA sequences (D2–D3 expansion segments of 28S rRNA gene and partial ITS1) and mtDNA sequences (partial COI mtDNA gene) from all known dagger nematode species and complete 16S rRNA from all endosymbiotic bacteria found in this survey (Table 1), together with other available selected sequences from the National Center for Biotechnology Information (NCBI) were used for phylogenetic analyses. Outgroup taxa for each studied gene was chosen according to previously published data by Archidona-Yuste et al. (2016a, b), Mobasseri et al. (2019a, b) and Palomares-Rius et al. (2016). MAFFT v. 7 (Katoh et al., 2019) and ClustalX2 (Larkin et al., 2007) with default parameters used for aligning. After sequence alignment, ambiguous regions were removed with Gblocks v. 0.91b (Castresana, 2000) using less stringent parameters (www.phylogeny.fr). Homogeneities of base frequencies and optional substitution models for 28S rRNA, ITS1, 16S rRNA, and COI mtRNA datasets were tested with Kakusan4 (Tanabe, 2011). Bayesian inference (BI) was performed using the software MrBayes v. 3.2.1 (Ronquist et al., 2012). For BI, the substitution model was selected by the Bayesian information criterion (BIC) and five different models were selected for our study. Convergence of the MCMC chains and burn-in length were assessed and checked in Tracer 1.7.1 (Rambaut et al., 2018). A number of 5,000,000 generations was performed for each BI analysis, sampling every 100th tree and discarding 25% of samples as ‘burn-in’. Finally, the final consensus trees were visualized using FigTree v. 1.4.3 (Rambaut, 2009).
Results
Morphology and morphometrics of dagger nematode species of the genus Xiphinema recovered from the West Coast of Syria
Syrian population of Xiphinema diffusum Lamberti & Bleve-Zacheo, 1979 (Fig. 1; Table 2).
Light micrographs of Xiphinema diffusum Lamberti & Bleve-Zacheo, 1979 females from the rhizosphere of banana (Musa sp.) from Hraisson (Baniyas, Tartous), Syria (A-J). A-B: Female anterior body region; C-D: Lip region; E: Basal bulb; F: Entire female specimen; G: Vulva region; H: J4 tail region; I: Female tail region; J: Bacterial endosymbionts. Abbreviations: a = anus; be = bacterial endosymbionts; gr = guiding ring; v = vulva. (Scale bars: A-C = 75 μm; D, H = 25 μm; E, I = 30 μm; F = 85 μm; G = 20 μm; J = 43 μm)
Distribution
Collected from the rhizosphere of banana (Musa sp.) trees at Hraisson, Baniyas district, Tartous, Syria (Table 1).
Brief description
The Syrian population of this species was characterised by a robust body, forming a close C-to open spiral when killed by heat. Cuticle appearing smooth, 2.5 (2.4–2.6) μm thick along body but thicker at the tail tip. Lip region expanded, rounded laterally and slightly flattened frontally, separated from the rest of the body by a shallow constriction. Odontostyle robust 1.7 (1.6–1.8) times longer than the odontophore, the latter with well-developed flanges (Fig. 1). Guiding sheath typical of the genus with the guiding ring at about 75.8 (71.4–79.1) μm distance from anterior end. Basal bulb cylindrical, 4.0 (3.5–4.7) times longer than wide. Glandularium 3.6 (3.4–3.8) times longer than basal bulb width. Cardia with hemispherical shape, 4.3 (3.9–4.5) μm long. Prerectum often indistinct. Rectum clearly visible 27 (24.4–29.1) μm long. Reproductive system didelphic-amphidelphic with equally developed branches, anterior branch 229 (213.2–246.5) μm long and posterior branch 258 (251.4–267.0) μm long, opposed, ovaries reflexed, and uteri short, without any differentiations. Ovaries showing bacterial endosymbionts (Fig. 1J). Vulva slightly post-equatorial, a transverse slit; vagina occupying two-third of the corresponding body width. Tail short, dorsally convex, slightly concave ventrally with rounded terminus. Males were not fround for this species. Only fourth (J4) juvenile stages were found and they were distinguished from other stages according to Robbins et al. (1996) (Table 2). Fourth (J4) juvenile stages were similar to adult specimens, except for their smaller size, longer tail, higher index c´, and absence of sexual characteristics (Fig. 1H).
Remarks
This species was found in a soil sample of banana (Musa sp.) trees (sample code: M64) at Hraisson, Baniyas district, Tartous, Syria (Table 1). The population presented low numbers of individuals in soil (20 individuals/500 cm3 of soil). The morphological and morphometric traits of this population closely agree with those of the type population (Lamberti & Bleve-Zacheo, 1979) and other subsequent records of this species (Chen et al., 2005; Heyns & Coomans, 1994; Lamberti et al., 1991, 2002; Lazarova et al., 2006; Luc et al., 1998; Shokoohi et al., 2021; Wang et al., 1996), except for minor intraspecific variations in the a and c ratios (e.g. 46–51 μm and 63–84 μm in the type population vs 33–41 μm and 56–60 μm), which may be because of geographical intraspecific variability. Apparently, our population of this species is almost indistinguishable from other species belonging to the X. brevicolle species complex (Lazarova et al., 2019), except by minor differences in morphological data, morphometric measures, but conspicuous differences in DNA sequences at level of the nematode and its associated bacterial endosymbionts. This species was originally described infecting Pelargonium plants in Réunion Island, Africa (Lamberti & Bleve-Zacheo, 1979) and later was reported from cultivated and wild ecosystems worldwide (Lamberti et al., 2000, 2002; Chen et al., 2005; Oliveira et al., 2005; Lazarova et al., 2006, 2019; Gutiérrez-Gutiérrez et al., 2016; Shokoohi et al., 2021). To our knowledge, these findings represent the first record for this species associated with banana plants in Syria, extending its geographical distribution in Asia.
The Syrian population of Xiphinema pachtaicum (Tulaganov, 1938) Kirjanova, 1951 (Fig. 2; Table 2).
Light micrographs of X. simile Lamberti et al., 1983 (A-B, D, F) and X. pachtaicum (Tulaganov, 1938) Kirjanova, 1951 (C, E) from the rhizosphere of banana (Musa sp.) from Hraisson (Baniyas, Tartous), and Siyannu (Jableh, Latakia), Syria (A-F). A: Female anterior body region; B-C: Lip region; D-E: Female tail region; F: Bacterial endosymbionts. Abbreviations: a = anus; be = bacterial endosymbionts. (Scale bars: A, E = 18 μm; B-C = 9 μm; D = 17 μm; F = 28 μm)
Distribution
Collected from the rhizosphere of banana (Musa sp.) trees at Siyannu, Jableh district, Latakia, Syria (Table 1).
Brief description
The Syrian population of this species was characterised by a slender body, forming a close C to coiled spiral, when killed by heat. Cuticle appearing smooth, 1.7 (1.5–1.9) μm thick along body but thicker at tail tip. Lip region expanded and distinctly offset from the rest of the body. Odontostyle long, 1.8 times longer than the odontophore with weak flanges. Guiding sheath typical of the genus with the guiding ring near the base of the odontostyle. Basal bulb cylindrical, 5.8 (5.5–6.0) times longer than wide. Glandularium 5.0 (4.8–5.2) times longer than basal bulb width. Cardia conoid-rounded, 4.2 (3.0–5.0) μm long. Prerectum indistinct. Rectum visible, 19.6 (18.7–21.0) μm long. Reproductive system didelphic-amphidelphic with two short and equally developed genital branches, anterior branch 196.1 (188.6–203.9) μm long and posterior branch 170.6 (162.0–179.7) μm long, ovaries opposed and reflexed, and uteri without any differentiations. Ovaries darkened in fixed specimen, endosymbionts apparently absent under light microscope. Vulva, posterior to mid-body, a transverse slit; vagina occupying two-third of the corresponding body width. Tail conoid with pointed terminus. Males and juveniles were not found for this species.
Remarks
This species was found in a soil sample of banana (Musa sp.) tree (sample code: M54) at Siyannu, Jableh district, Latakia, Syria (Table 1). This population presented low specimens densitiy (14 individuals/500 cm3 of soil). The morphological traits and morphometric data of females closely agree with those of the described population by Lamberti and Siddiqi (1977) and with other populations already reported (Barsi & Lamberti, 2002; Fadaei et al., 2003; Getaneh et al., 2015; Gutiérrez-Gutiérrez et al., 2011a, b; Kumari et al., 2005; Lamberti & Bleve-Zacheo, 1979; Lamberti et al., 1993; Mobasseri et al., 2019a, b; Vazifeh et al., 2019). Our population of this species is almost indistinguishable from other X. pachtaicum-subgroup species (Gutiérrez-Gutiérrez et al., 2012; Archidona-Yuste et al., 2016b), except by some indiscernible morpho-anatomical differences, but notable differences in DNA sequences. Although this species has already been reported from Syria by Lamberti (1984) and Greco et al. (1992), no molecular and morphometric data were formerly provided for those populations. This species is widespread in the countries bordering the Mediterranean Sea, central and Eastern Europe, Africa and Asia (Gutiérrez-Gutiérrez et al., 2016; Lamberti et al., 2000, 2002; Taylor & Brown, 1997). To our knowledge, this is the first record of this species associated with banana plants in Syria. Furthermore, these findings confirm that it is widely distributed in the Middle Eastern.
The Syrian population of Xiphinema simile Lamberti et al., 1983 (Fig. 2; Table 2).
Distribution
Collected from the rhizosphere of banana (Musa sp.) trees at Hraisson, Baniyas district, Tartous, Syria (Table 1).
Brief description
The Syrian population of this species was characterised by a slender body, forming a close C to spiral shape when killed by heat. Cuticle smooth under light microscope, 1.4 (1.3–1.5) μm thick along body but thicker at tail tip. Lip region expanded, flat, rounded in corners, and offset from the rest of the body. Odontostyle 1.8 (1.8–1.9) times longer than the odontophore, the latter with weak flanges. Guiding ring 62.7 (61.9–63.4) μm from the anterior end. Basal bulb cylindrical, 6.4 (5.6–7.4) times longer than wide. Glandularium 5.4 (5.0–5.7) times longer than basal bulb width. Cardia conoid-rounded shape, 3.2 (2.8–3.6) μm long. Prerectum indistinct. Rectum 16.7 (14.5–18.0) μm long. Reproductive system didelphic-amphidelphic with two equally developed branches, anterior branch 165.3 (153.0–188.1) μm long and posterior branch 154.6 (150.1–160.2) μm long, opposed and reflexed ovaries with bacterial symbionts, and short uteri without any differentiation, vulva, posterior to mid-body, a transverse slit; vagina occupying 55–59% of the corresponding body diameter. Tail conical, dorsally convex, with finely rounded terminus. Males and juveniles were not found for this species.
Remarks
This species was found in a soil sample of banana (Musa sp.) tree (sample code: M69) at Hraisson, Baniyas district, Tartous, Syria (Table 1) in low density (18 individuals/500 cm3 of soil). The morphological features and morphometric measurements of females closely agree with those of the type population (Lamberti et al., 1983) and with other populations from Europe (Barsi & De Luca, 2008; Barsi & Lamberti, 2002, 2004; Barsi, 1994a; Kumari, 2006; Lamberti et al., 1999b; Lazarova et al., 2008; Lišková, 1995; Lišková & Brown, 1996; Lišková et al., 1993; Peneva & Choleva, 1992; Repasi et al., 2008) and outside Europe (Coomans & Heyns, 1997; Naghavi et al., 2022a). It is morphologically close to other members of X. simile-subgroup species (Lazarova et al., 2016a, b), except by some minor morphological and morphometric differences, but remarkable differences in DNA sequences. Xiphinema simile is widespread in the European countries in association with agricultural crops, and landscape trees and grasslands (Barsi & De Luca, 2008; Barsi & Lamberti, 2002, 2004; Barsi, 1994a; Kumari, 2006; Lamberti et al., 1999b, 2000; Lazarova et al., 2008; Lišková, 1995; Lišková & Brown, 1996; Lišková et al., 1993; Peneva & Choleva, 1992; Repasi et al., 2008; Taylor & Brown, 1997). Outside Europe, this species has been only reported from Kenya (Coomans & Heyns, 1997) and Iran (Naghavi et al., 2022a). To our knowledge this is the first record of this species infesting soils around banana trees in Syria, extending its geographical distribution in the Middle Eastern.
The Syrian populations of Xiphinema vuittenezi Luc, Lima, Weischer, Flegg, 1964 (Fig. 3; Table 2).
Light micrographs of Xiphinema vuittenezi Luc, Lima, Weischer, Flegg, 1964 females from the rhizosphere of banana (Musa sp.) from Hraisson (Baniyas, Tartous), and Siyannu (Jableh, Lattakia), Syria (A-I). A: Lip region; B: Female tail region; C: J4 tail; D: J3 tail; E: Posterior reproductive branch; F: Vulva region; G-I: detail of female genital track showing crystalloid bodies. Abbreviations: a = anus; cb = crystal bodies; gr = guiding ring; ov = ovary; v = vulva. (Scale bars: A = 13 μm; B = 41 μm; C-D = 30 μm; E: 58 μm; F-I = 29 μm)
Distribution
It was recovered from two soil samples collected from the rhizosphere of banana (Musa sp.) trees at Hraisson, Baniyas district, Tartous, Syria (Table 1).
Brief description
The Syrian population of this species was characterised by a cylindrical body, forming a close C, when killed by heat. Cuticle smooth under light microscope, 3.3 (2.6–3.8) μm thick along body but thicker at tail tip. Lip region rounded, offset from the rest of the body by depression. Odontostyle robust, two times longer than the odontophore, the later with well-developed flanges. Guiding ring 126.7 (114.9–133.0) μm from the anterior end. Basal bulb cylindrical, 4.8 (4.0–5.9) times longer than wide. Glandularium 4.2 (3.6–4.9) times longer than basal bulb width. Cardia conoid-rounded, 6.2 (5.8–6.5) μm long. Prerectum 579.5 (573.7–585.3) μm long. Rectum as long as the anal body diameter. Reproductive system didelphic-amphidelphic with branches about equally developed without any Z-differentiation but containing crystalloid bodies distributed over the entire length (Fig. 3). Vulva a transverse slit, situated at mid-body; vagina usually occupying half of the corresponding body diameter. Tail short, broadly round, with a peg (Fig. 3). Males were not found for this species. Third (J3) and fourth (J4) juvenile stages were found and they were similar to adult specimens, except for their smaller size, higher index c´, and absence of sexual characteristics (Fig. 3). All stages are distinguishable by relative body lengths, functional, and replacement odontostyle (Robbins et al., 1996) (Table 2). J3 characterized by a conical tail with a rounded end subdigitate extension (Fig. 3). J4 characterized by a tail bluntly rounded, slightly dorsally convex-conoid with a terminal peg (Fig. 3).
Remarks
This species was found in two soil samples collected from the rhizosphere of banana (Musa sp.) trees (sample codes: M78 and M79) at Hraisson, Baniyas district, Tartous, Syria (Table 1). The only one adult specimen that was detected in the sample M78 was checked under light microscope before sequencing it. Then, this population (M78) was only molecularly characterized. The code 79 showed low nematode density (14 individuals/500 cm3 of soil). The morphological features and morphometric measurements of the fixed females closely agree with those of the type population (Luc et al., 1964), in which their authors did not consider the presence of crystalloids in this species (Coomans et al., 2001). It matches well with other populations from Europe and Middle East (i.e. Mojtahedi et al., 1980; Barsi, 1989, 1994b; Roca et al., 1989, 1991; Coiro et al., 1989, 1992; Barsi & Lamberti, 2000; Kumari et al., 2005; Groza et al., 2013; Gutiérrez-Gutiérrez et al., 2016; Vazifeh et al., 2019; Naghavi et al., 2022b), except for minor differences in the body length and b ratio (e.g. 2800–3400 μm and 6.7–7.0; 3008–4109 μm and 6.0–8.7; 3250 –3760 μm and 7.3–7.9 vs 2369–2739 μm, 4.0–6.5) (Kumari et al., 2005; Roca et al., 1989; Vazifeh et al., 2019) which may be because of geographical intraspecific variability. This species is widespread in the European countries, especially Central Europe (Mojtahedi et al., 1980; Lamberti et al., 1983; Barsi, 1989, 1994b; Roca et al., 1989, 1991; Taylor & Brown, 1997; Coiro et al., 1989, 1992; Barsi & Lamberti, 2000; Coomans et al., 2001; Walker, 2004; Kumari et al., 2005; Groza et al., 2013; Vazifeh et al., 2019; Naghavi et al., 2022b). To our knowledge this is the first record of this species associated with banana plants. Furthermore, these findings represent the first detection of this species in Syria.
The Syrian population of Xiphinema zagrosense Ghaemi, Pourjam, Pedram, Robbins, Ye & Decraemer, 2012 (Fig. 4; Table 2).
Light micrographs of Xiphinema zagrosense Ghaemi, Pourjam, Pedram, Robbins, Ye & Decraemer, 2012 female from the rhizosphere of banana (Musa sp.) from Hraisson (Baniyas, Tartous), Syria (A-H). A-B: Lip regions; C: Vulva region; D: detail of female genital track showing crystalloid bodies; E–F: Female tail region; G: Detail of basal bulb; H: posterior uterine branch. Abbreviations: a = anus; cb = crystalloid bodies; ov = ovary; v = vulva. (Scale bars: A-B = 22 μm; C-D = 35 μm; E–F: 48 μm; G = 42 μm; H: 70 μm)
Distribution
Collected from the rhizosphere of banana (Musa sp.) trees at Hraisson, Baniyas district, Tartous, Syria (Table 1).
Brief description
The Syrian population of this species is characterised by a cylindrical body, forming an open C shape when killed by heat. Cuticle smooth under light microscope, 3.57 μm thick at mid-body. Lip region rounded, separate from the rest of the body by a slight depression. Odontostyle robust, 1.5 times longer than the odontophore, the later with well-developed flanges. Guiding ring 157.2 μm from the anterior end. Basal bulb cylindrical, eight times longer than wide. Glandularium seven times longer than basal bulb width. Cardia semi-circular, 7.1 μm long. Prerectum 11 times the anal body diameter and rectum as long as the anal body diameter. Reproductive system didelphic-amphidelphic with branches about equally developed without Z-differentiation, uterus bipartite which is composed of pars dilatata uteri and tubular portion with crystalloids (Fig. 4). Vulva a transverse slit, situated at mid-body; vagina occupying more than half of the corresponding body diameter. Tail convex conoid with a broadly rounded terminus (Fig. 4). Male and juveniles were not found for this species.
Remarks
This dagger nematode species was found in a soil sample of banana (Musa sp.) tree (sample codes: M74) at Hraisson, Baniyas district, Tartous, Syria (Table 1). It showed a low density (2 individuals/500 cm3 of soil). The morphological and morphometric features of females closely agree with those of the type population (Ghaemi et al., 2012), except for minor differences in the lip region diameter and height (15.0–18.0 μm and 7.0–9.0 μm vs 13.6 μm and 4.6 μm), which should be considered as an intraspecific variability. This species is morphologically close to members of X. pyrenaicum complex (Gutiérrez-Gutiérrez et al., 2010; Lamberti et al., 1992; Pedram et al., 2009, 2012), except by some indiscernible morpho-anatomical differences, but notable differences in DNA sequences. This species was described in association with grasses in natural environments from Iran (Ghaemi et al., 2012). To our knowledge, this is the first record of this species outside its type locality, extending its distribution in the world, and first report of its association with banana plants.
Sequence analysis and phylogeny of 28S rRNA gene
Xiphinema americanum-group spp.
The two 28S sequences of the Syrian isolate of X. diffusum (OR196832, OR196837) obtained showed high homology (99–100%) with X. brevicolle complex, such as X. diffusum (e.g., AY601600, MK050001, and MK050002) and X. brevicolle Lordello & Da Costa 1961 (e.g., KY512785– KY512787 and AY601601), but presented less than 98% homology with the topotype population of X. brevicolle (MH248813). The 28S sequence of X. simile (OR196834) showed a high homology (99–100%) with sequences of the same species, namely X. simile (AY601609, JQ780368–JQ780370, KJ802889–KJ802890, ON500623, MK957227–MK957228). For these species, the respective Bayesian phylogenetic tree (Fig. 5) showed that X. diffusum and X. brevicolle were grouped together within the well-supported clade of these same species and clearly separated from the topotype population of X. brevicolle (MH248813). These results could be due to a possible misidentification of the species in GenBank. Although the Syrian isolate of X. simile was phylogenetically closest to European and Asian populations of the same species (e.g. AY601609, JQ780368–JQ780370, KJ802889– KJ802890), the length of all these sequences were different, yielding intraspecies cladogenesis and varied branch lengths inside this species clade.
Phylogenetic relationships of Xiphinema diffusum Lamberti & Bleve-Zacheo, 1979 and X. simile Lamberti et al., 1983. Bayesian 50% majority rule consensus trees was inferred from 96 sequences of D2-D3 expansion segments (713 bp) of 28S rRNA sequences aligned under the SYM + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are coloured in red and navy blue. Scale bar = expected changes per site
Xiphinema non americanum-group spp.
The 28S sequences from Syrian populations of X. vuittenezi (OR196835–OR196836, OR428495–OR428496) matched closely (more than 99% of similarity) to an Iran poputation of the same species (MK905505), varying in 4–13 nucleotides and 0–5 indels. Our sequences matched also with other world isolates with 98–99% similarity (e.g. HG329724–HG329724, HG969310, AY601614, MK943733, MK957234, EF614266, MK957237, MK957239). Little intra-specific variation between the 28S sequences from the two Syrian populations of X. vuittenezi from Hraisson (M78 and M79, banana) was recorded (0–2 nucleotides and 0–1 indel) (Table 1). The 28S sequence of X. zagrosense (OR196833) showed high homology (99–100%) with X. zagrosense (JN153101, Iran population) with low sequence variations (only 4 nucleotides with no indels). The Bayesian phylogenetic tree of X. vuittenezi and X. zagrosense is presented in Fig. 6. The Syrian isolates of X. vuittenezi were closest to isolates of the same species from different world populations but not within the same clade (Fig. 6), mostly due to the intraspecific variations of the 28S sequences from isolates that distinguished Hungarian, Iranian, Romanian, Czech and Russian populations from the Syrian, yielding intraspecies cladogenesis with two subclades inside this species clade. In relation to X. zagrosense, the Iranian (JN153101) and Syrian (OR196833) populations clustered in a well-supported sub-clade.
Phylogenetic relationships of Xiphinema vuittenezi Luc et al., 1964, and X. zagrosense Ghaemi, Pourjam, Pedram, Robbins, Ye, & Decraemer, 2012. Bayesian 50% majority rule consensus trees was inferred for 143 sequences of D2-D3 expansion segments (673 bp) of 28S rRNA sequences aligned under the GTR + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are coloured in green and pink. Scale bar = expected changes per site
Sequence analysis and phylogeny of ITS rRNA
Xiphinema americanum-group spp.
Concerning the ITS region, the Syrian population of X. diffusum (OR197584) presented 95–96% sequence similarity with other X. brevicolle complex sequences, such as X. diffusum (e.g., AY359858, MK050006) and X. brevicolle (e.g., FM211423, FM211420, FM211426, HQ184474, KY512780), with a considerable intra-species variability recording 21–48 nucleotide differences and 8–13 indels. For X. pachtaicum, the Syrian ITS sequence (OR197587) showed more than 99% match with other sequences from the same species (e.g. AY430178, KU250149), presenting little sequence polymorphisms (0–5 nucleotides and 0–3 indels). The ITS sequence of X. simile (OR264513) showed 95% sequence similarity with the Spanish X. simile isolates (KX263242– KX263243), followed by Iranian isolates of X. persicum Jahanshahi Afshar et al. (2021) (94% of similarity) (MT073075, MT073077– MT073078). Our ITS sequences of X. simile showed an intraspecific variability with 30–29 nucleotides and 10–11 indels. As for their phylogenetic inference (Fig. 7), the Syrian populations of X. diffusum (OR197584) were closelly related with several isolates of the X. brevicolle complex from different countries and clearly separated from the topotype population of X. brevicolle (MH248804). All ITS sequences of X. diffusum clustered together within the same clade; however, Korean and Syrian isolates (e.i. OR197584, MK050006) were separated from the South African isolate (AY359858). This could be dues to X. diffusum has variation among some isolates for this marker. The ITS sequences of X. pachtaicum and X. incertum Lamberti et al., 1983 were clustered in the same clade, mostly because the sequences were shorter and poor quality in the database. The ITS sequences of X. simile and X. persicum were also grouped in the same clade, probably due to the high intraspecific variation among some populations.
Phylogenetic relationships of Xiphinema diffusum Lamberti & Bleve-Zacheo, 1979, X. simile Lamberti et al., 1983, and X. pachtaicum (Tulaganov, 1938) Kirjanova, 1951. Bayesian 50% majority rule consensus trees was inferred from 61 sequences of ITS1 rRNA (381 bp) aligned under the K80 + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are coloured in red, navy blue, and sky blue. Scale bar = expected changes per site
Xiphinema non americanum-group spp.
The two ITS sequences from Syrian populations of X. vuittenezi (OR197585– OR197586), that shared in-between 99% similarity, showed 89–91% sequence similarity with several isolates of X. vuittenezi (e.g. HG329722–HG329723, HG969308–HG969308; MK909917; AJ437028) and 89% similarity with X. iranicum Pedram et al., 2009 (EU477386); presenting a intraspecific variation of 3–46 nucleotide mismatches and 0–15 indels. The corresponding Bayesian phylogenetic tree is represented in Fig. 8. The Syrian ITS sequences of X. vuittenezi clustered together in a well-supported subclade within the clade of X. vuittenezi (e.i. OR197585– OR197585, AY359858, HG969308– HG969309, MK909917, HG329722–HG329723, AJ437028) and X. iranicum (EU477386), probably due to X. vuittenezi shows a high intraspecific variation for this marker as well as the shorter and poor quality in some sequences deposited in the database.
Phylogenetic relationships of Xiphinema vuittenezi Luc et al., 1964. Bayesian 50% majority rule consensus trees was inferred from 61 sequences of ITS1 rRNA (260 bp) aligned under the K80 + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are coloured in green. Scale bar = expected changes per site
Sequence analysis and phylogeny of COI mtDNA
Xiphinema americanum-group spp.
The Syrian COI sequences of X. diffusum (OR196695) showed a sequence similarity of 97–100% to sequences from the X. brevicolle complex, such as X. diffusum (e.g., AM086698– AM086701) and X. brevicolle (e.g., ON107524, ON107526– ON107529), with intraspecific variation of 1–9 nucleotides. For X. pachtaicum, our COI sequence (OR196696) showed high homology (98–100% of similarity) with other sequences of this species (e.g. KX263112–KX263113, HM921378), with 0–8 nucleotides mismatches. Regarding the phylogenetic tree (Fig. 9), the Syrian COI sequences of X. diffusum and X. brevicolle grouped together in the well-supported clade and clearly separated from the topotype population of X. brevicolle (AM086707, MH248818). This could be dues to a plausible misidentification of these species for some sequences available in GenBank. Moreover, all COI sequences of X. pachtaicum also clustered in a well-supported sub-clade, however, the length of some sequences was different, yielding different branch lengths inside this species clade.
Phylogenetic relationships of Xiphinema diffusum Lamberti & Bleve-Zacheo, 1979 and X. simile Lamberti et al., 1983 and X. pachtaicum (Tulaganov, 1938) Kirjanova, 1951. Bayesian 50% majority rule consensus trees was inferred from 91 sequences of COI mtDNA (341 bp) aligned under the HKY85 + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are coloured in red and sky blue. Scale bar = expected changes per site
Xiphinema non americanum-group spp.
The COI sequence from Syrian population of X. vuittenezi (OR196697) showed 83% similarity to the Russian isolate of this species (OQ699231) and less than 79% similarity with other Xiphinema species. Regarding the phylogenetic inference (Fig. 10) showed that our X. vuittenezi COI sequence was clustered, with good support, in-between X. afratakhtehense (MH429098) and the cluster of the Russian and Czech isolates of X. vuittenezi (OQ699231 and EF614265). This could be dues to a poor quality of these sequences and overlapping peaks, yielding intraspecies cladogenesis.
Phylogenetic relationships of Xiphinema vuittenezi Luc et al., 1964. Bayesian 50% majority rule consensus trees was inferred from 65 sequences of COI mtDNA (335 bp) sequences aligned under the GTR + G + I model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are coloured in green. Scale bar = expected changes per site
Phylogenetic analysis of the bacterial endosymbionts 'Candidatus Xiphinematobacter’ and correlation with host phylogeny
The complete 16S rRNA gene of the bacterial endosymbionts 'Candidatus Xiphinematobacter' were successfully amplified from X. diffusum (OR196969, OR196971) and X. simile (OR196970), with an approximate size of 1400 bp. Both sequences from X. diffusum were 100% identical and matched closely (99–100% identity) with several sequences of Chinese, Brazilian, and Australian Xiphinematobacter species reported from X. diffusum populations (KJ614450–KJ614453). A sequence of South African population of Xiphinematobacter from X. brevicolle complex (AF217462) also grouped in this cluster. When compared with Xiphinematobacter species detected in topotype population of X. brevicolle (KJ614447), the latter sequence had only 96% identity. The other endosymbiont from X. simile (OR196970) matched closely (99–100% identity) with two sequences of a Greek population of this same species (KT735097–KT735098) from X. simile. Figure 11 presents the BI tree (50% majority rule consensus tree) of 70 sequences of 16S rRNA for ‘Ca. Xiphinematobacter’ and three outgroup species [Uncultured Eubacterium spp. WCHD3-88 (AF050561), Spartobacteria bacterium (AB245341) and Terrimicrobium sacchariphilum (GU129926)]. The phylogenetic analysis within the genus ‘Candidatus Xiphinematobacter’ differentiated 18 well-supported clades corresponding to the 18 species denoted as species A through species R (Fig. 11). In exception of ‘Candidatus Xiphinematobacter’ sp. A, which was isolated from two X. americanum s.s., X. pachtaicum, and X. simile from Europe (KT735098, KJ614457, KJ614458) and Asia (MT568602, OR196970, KX263187), and ‘Candidatus Xiphinematobacter’ sp. R, which was isolated from X. americanum s. str., X. americanum s. s., X. rivesi Dalmaso, 1969, X. peruvianum Lamberti & Bleve-Zacheo, 1979, X. georgianum Lamberti & Bleve-Zacheo, 1979, the other ‘Candidatus Xiphinematobacter’ species were all isolated from one or two host nematode species. ‘Candidatus Xiphinematobacter’ sp. I included two sequences of one population of X. diffusum from Syria, which grouped in the well-supported clade with other populations from Asia, Australia, Africa, and South America, and one population of X. brevicolle complex from South Africa, with high clade support (0.95 BPP), clearly separated of the sequence of ‘Candidatus Xiphinematobacter’ sp. G of the topotype population of X. brevicolle. ‘Candidatus Xiphinematobacter’ from X. simile from Syria clustered in the sub-clade of ‘Candidatus Xiphinematobacter’ sp. A, jointly with other sequences of the bacterium from populations of X. simile from other origins.
Phylogenetic relationships of the endosymbiont bacterias, isolated from Xiphinema diffusum Lamberti & Bleve-Zacheo, 1979 and X. simile Lamberti et al., 1983 within ‘Candidatus Xiphinematobacter’. Bayesian 50% majority rule consensus tree was inferred from 16S rRNA sequence alignment under the K80 + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are targed with a arrow. Scale bar = expected changes per site
Discussion
In this study, six Xiphinema populations were recovered and identified from Syria belonging to five species, namely X. diffusum, X. pachtaicum, X. simile, X. vuittenezi and X. zagrosense by integrating morphological and molecular data. Using an integrative approach to identify species in this specious genus has already well documented (Gutiérrez-Gutiérrez et al., 2012; Ghaemi et al., 2012; Archidona-Yuste et al., 2016b, 2020a; Lazarova et al., 2019; Mobasseri et al., 2019a, b; Vazifeh et al., 2019; Naghavi et al., 2022a; Gu et al., 2022). The endosymbiont bacterias were also sequenced for two X. americanum-group species, X. diffusum and X. simile. Four species including X. diffusum, X. simile, X. vuittenezi, and X. zagrosense are new records from Syria, increasing the geographical distribution points of these species.
In accordance with other taxonomic studies (Lazarova et al., 2008, 2016b; Gutiérrez-Gutiérrez et al., 2011b; 2013; Archidona-Yuste et al., 2016a, b; 2020a, b; Mobasseri et al., 2019a, b; Vazifeh et al., 2019; Naghavi et al., 2022a), our results showed that the D2–D3 of the 28S rRNA and the COI mtDNA regions were the most conclusive molecular markers for species-level identification within genus Xiphinema in exception of some species within the subgroup X. brevicolle. Our study corroborates other studies (Lazarova et al., 2016a; Mobasseri et al., 2019a, b; Palomares-Rius et al., 2016) emphasizhing that 16S rRNA gene is a useful molecular marker for discerning among Candidatus Xiphinematobacter species and so, as a result of their closely related Xiphinema hosts. In our phylogenetic, representative sequences for some species occupied different positions. An accurate way to resolve this problem is to sequence topotype specimens. Currently, most populations of X. brevicolle have identified only based on molecular data without morphological and morphometric analysis, occupying distant placements from the sequences of the topotype population of the species, and possibly indicating misidentification. Furthermore, recently Jahanshahi Afshar (2019) sequenced the D2–D3 fragments of 28S rRNA gene of the topotype population of X. robbinsi Pedram, Niknam & Decraemer, 2008 revealing a misktake identification of a Tunisian population.
Nowadays, exploring the molecular evolution and phylogenetic relationships of ‘Ca. Xiphinematobacter’ species using the 16S rRNA gene is at its peak (Brown, 2018; Lazarova et al., 2016a, b; Mobasseri et al., 2019a, b; Palomares-Rius et al., 2016, 2021). Still, there is knowledge gap on the nematode-endosymbiont bacteria co-evolution in the X. americanum group. Our results agree with other authors (Mobasseri et al., 2019a, b; Palomares-Rius et al., 2016, 2021) that proposed a plausible host-endosymbiont co-speciation and suggest an adaptative role during the process of natural selection in the host Xiphinema species.
In conclusion, our work revealed a high biodiversity of the dagger nematode species in banana gardens of Syria. The recovered species could be regarded potentioal threats and monitoring of their population could prevent their outbreak.
References
Abdine, M., Allak, H., Nus, B. E., Nigro, F., Catalano, L., & Digiaro, M. (2007). Phytosanitary Aspects and Nursery Production of Olive in Syria: pitfalls and perspectives. In B. Di Terlizzi, A. Dragotta, & M. Jamal (Eds.), Syrian national strategic plan for olive oil quality. Final report, Bari: CIHEAM, Options Méditerranéennes: Série A. Séminaires Méditerranéens; n. 73 (pp. 143–151)
Ahmed, M., & Holovachov, O. (2021). Twenty years after De Ley & Blaxter – how far did we progress in understanding the phylogeny of the phylum Nematoda? Animals, 11(12), 3479. https://doi.org/10.3390/ani11123479
Al-Halabi, S. M., & Al-Assas, K. (2021). Survey of parasitic nematode genera associated with grapevine roots in Sweida governorate. Syria. Arab Journal of Plant Protection, 39(1), 14–21. https://doi.org/10.22268/AJPP-39.1.014021[inArabic]
Ali, N. (2023). Incidence and diversity of plant parasitic nematode communities associated with greenhouse ornamental plants in the coastal region of Syria. European Journal of Plant Pathology, 166, 491–508. https://doi.org/10.1007/s10658-023-02678-z
Allouf, N. (2013). A preliminary survey of nematodes genera associated with Citrus Trees in Syrian Coast. The Jordanian Journal of Agricultural Sciences, 9(3), 395–403. [in Arabic].
Archidona-Yuste, A., Navas-Cortes, J. A., Cantalapiedra-Navarrete, C., Palomares-Rius, J. E., & Castillo, P. (2016a). Remarkable diversity and prevalence of dagger nematodes of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) in olives revealed by integrative approaches. PLoS ONE, 11(11), e0165412. https://doi.org/10.1371/journal.pone.0165412
Archidona-Yuste, A., Navas-Cortes, J. A., Cantalapiedra-Navarrete, C., Palomares-Rius, J. E., & Castillo, P. (2016b). Cryptic diversity and species delimitation in the Xiphinema americanum-group complex (Nematoda: Longidoridae) as inferred from morphometrics and molecular markers. Zoological Journal of the Linnean Society, 176(2), 231–265. https://doi.org/10.1111/zoj.12316
Archidona-Yuste, A., Wiegand, T., Castillo, P., & Navas-Cortés, J. A. (2019). Dataset on the diversity of plant-parasitic nematodes in cultivated olive trees in southern Spain. Data in Brief, 27, 104658. https://doi.org/10.1016/j.dib.2019.104658
Archidona-Yuste, A., Wiegand, T., Castillo, P., & Navas-Cortes, J. A. (2020a). Spatial structure and soil properties shape local community structure of plant-parasitic nematodes in cultivated olive trees in southern Spain. Agriculture, Ecosystems & Environment, 287, 106688. https://doi.org/10.1016/j.agee.2019.106688
Archidona-Yuste, A., Cai, R., Cantalapiedra-Navarrete, C., Carreira, J. A., Rey, A., Viñegla, B., Liébanas, G., Palomares-Rius, J. E., & Castillo, P. (2020b). Morphostatic speciation within the dagger nematode Xiphinema hispanum-Complex species (Nematoda: Longidoridae). Plants, 9(12), 1649. https://doi.org/10.3390/plants912164
Barsi, L. (1989). The Longidoridae (Nematoda: Dorylaimida) in Yugoslavia. I. Nematologia Mediterranea, 17, 97–108.
Barsi, L. (1994a). Species of the Xiphinema americanum-group (Nematoda: Dorylaimida) on the territory of the former Yugoslavia. Nematologia Mediterranea, 22(1), 25–34.
Barsi, L. (1994b). Bivulval females of Longidorus euonymus, Xiphinema diversicaudatum and X. vuittenezi (Nematoda: Dorylaimida). Nematologia Mediterranea, 22, 271–272.
Barsi, L., & De Luca, F. (2008). Morphological and molecular characterisation of two putative Xiphinema americanum-group species, X. parasimile and X. simile (Nematoda: Dorylaimida) from Serbia. Nematology, 10, 15–25. https://doi.org/10.1163/156854108783360212
Barsi, L., & Lamberti, F. (2000). Morphometric variation and juvenile stages of Xiphinema vuittenezi (Nematoda: Dorylaimida) in Serbia. Nematologia Mediterranea, 28, 3–12.
Barsi, L., & Lamberti, F. (2002). Morphometrics of three putative species of the Xiphinema americanum-group (Nematoda: Dorylaimida) from the territory of the former Yugoslavia. Nematologia Mediterranea, 30, 59–72.
Barsi, L., & Lamberti, F. (2004). Xiphinema parasimile sp. n. from Serbia and X. simile, first record from Bosnia and Herzegovina (Nematoda, Dorylaimida). Nematologia Mediterranea, 32(1), 101–109.
Brown, A. M. (2018). Endosymbionts of plant-parasitic nematodes. Annual Review of Phytopathology, 56, 225–242. https://doi.org/10.1146/annurev-phyto-080417-045824
Cai, R., Maria, M., Barsalote, E. M., Subbotin, S. A., & Zheng, J. (2018). Description of Xiphinema hangzhouense sp. n. (Nematoda: Longidoridae) from the rhizosphere of Magnolia grandiflora in Hangzhou, Zhejiang Province, China. Nematology, 20(1), 67–80. https://doi.org/10.1163/15685411-00003125
Cai, R., Archidona-Yuste, A., Cantalapiedra-Navarrete, C., Palomares-Rius, J. E., Zheng, J., & Castillo, P. (2019). Integrative taxonomy of Xiphinema histriae and Xiphinema lapidosum from Spain. Journal of Nematology, 51, 1–21. https://doi.org/10.21307/jofnem-2019-037
Cai, R., Archidona-Yuste, A., Cantalapiedra-Navarrete, C., Palomares-Rius, J. E., & Castillo, P. (2020). New evidence of cryptic speciation in the family Longidoridae (Nematoda: Dorylaimida). Journal of Zoological Systematics and Evolutionary Research, 58(4), 869–899.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17(4), 540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Chen, D. Y., Ni, H. F., Yen, J. H., Cheng, Y. H., & Tsay, T. T. (2005). Differentiation of the Xiphinema americanum-group nematodes X. brevicollum, X. incognitum, X. diffusum and X. oxycaudatum in Taiwan by morphometrics and nuclear ribosomal DNA sequences. Nematology, 7(5), 713–725.
Coiro, M. I., Lamberti, F., Agostinelli, A., & Vindimian, M. E. (1989). I Longidoridae nei vigneti del Trentino. II: Il genere Xiphinema Cobb. Nematologia Mediterranea, 17, 139–148.
Coiro, M. I., Agostinelli, A., & Lamberti, F. (1992). Longidoridae (Nematoda) in the vineyards of the province of Verona. Nematologia Mediterranea, 20, 87–95.
Coomans, A., & Heyns, J. (1997). Three species of the Xiphinema americanum-group (Nematoda: Longidoridae) from Kenya. Nematologica, 43, 259–274.
Coomans, A., Huys, R., Heyns, J., Luc, M. (2001). Character analysis, phylogeny and biogeography of the genus Xiphinema Cobb, 1913 (Nematoda, Longidoridae). Vol. 287. Annales du Musée Royal de l’Afrique Centrale (Zoologie), Tervuren, Belgique, 239 pp.
De Grisse, A. T. (1969). Redescription ou modifications de quelques techniques utilisées dans l’étude de nématodes phytoparasitaires. Mededelingen Rijksfaculteit Landbouwwetenschappen Gent, 34, 315–359.
de Man, J. G. (1880). Die einheimischen, frei in der reinen Erde und im su ¨ssen Wasser lebende Nematoden, monographischen bearbeitet. Tijdschrift Nederlandse Dierkundige vereniging. 104 p.
Fadaei, A. A., Coomans, A., & Kheiri, A. (2003). Three species of the Xiphinema americanum lineage (Nematoda: Longidoridae) from Iran. Nematology, 5, 453–461. https://doi.org/10.1163/156854103-769224430
Flegg, J. J. M. (1967). Extraction of Xiphinema and Longidorus species from soil by a modification of Cobb’s decanting and sieving technique. Annals of Applied Biology, 60(3), 429–437.
Fouladvand, Z. M., Pourjam, E., Castillo, P., & Pedram, M. (2019). Genetic diversity, and description of a new dagger nematode, Xiphinema afratakhtehnsis sp. nov., (Dorylaimida: Longidoridae) in natural forests of southeastern Gorgan, Northern Iran. PloS one, 14(5), e0214147. https://doi.org/10.1371/journal.pone.0214147
Getaneh, G., Bert, W., & Decraemer, W. (2015). First report, morphological and molecular characterization of Xiphinema elongatum and X. pachtaicum (Nematoda, Longidoridae) from Ethiopia. Zookeys, 489, 1–13. https://doi.org/10.3897/zookeys.489.8629
Ghaemi, E., Pourjam, E., Pedram, M., Robbins, R. T., Ye, W., & Decraemer, W. (2012). Morphological and molecular characterisation of Xiphinema zagrosense sp. n. (Dorylaimida: Longidoridae) from the Zagros Mountains. Iran. Nematology, 14(4), 445–55.
Greco, N., Di Vito, M., Reddy, M. V., & Saxena, M. C. (1984). A preliminary report of survey of plant parasitic nematodes of leguminous crops in Syria. Nematologia Mediterranea, 12, 87–93.
Greco, N., Di Vito, M., & Saxena, M. C. (1992). Plant parasitic nematodes of cool season food legumes in Syria. Nematologia Mediterranea, 20(1), 37–46.
Groza, M., Lazarova, S., Costache, C., Luca, F., Rosca, I., Fanelli, E., & Peneva, V. (2013). Morphological characterisation and diagnostics of Xiphinema non-americanum group species (Nematoda: Longidoridae) from Romania using mutiplex PCR. Helminthologia, 50(3), 215–231. https://doi.org/10.2478/s11687-013-0133-3
Gu, J., Ye, W., & Munawar, M. (2022). Description of Xiphinema pupureum n. sp. (Nematoda: Longidoridae), a new Xiphinema americanum group species detected from the rhizosphere of Ilex purpurea from Japan. Nematology, 25(1), 91–105. https://doi.org/10.1163/15685411-bja10208
Gutiérrez-Gutiérrez, C., Palomares-Rius, J. E., Cantalapiedra-Navarrete, C., Landa, B. B., Esmenjaud, D., & Castillo, P. (2010). Molecular analysis and comparative morphology to resolve a complex of cryptic Xiphinema species. Zoologica Scripta, 39(5), 483–498. https://doi.org/10.1111/j.1463-6409.2010.00437.x
Gutiérrez-Gutiérrez, C., Castillo, P., Cantalapiedra-Navarrete, C., Landa, B. B., Derycke, S., & Palomares-Rius, J. E. (2011a). Genetic structure of Xiphinema pachtaicum and X. index populations based on mitochondrial DNA variation. Phytopathology, 101(10), 1168–1175.
Gutiérrez-Gutiérrez, C., Palomares Rius, J. E., Cantalapiedra-Navarrete, C., Landa, B. B., Esmenjaud, D., & Castillo, P. (2011b). Prevalence, polyphasic identification, and molecular phylogeny of dagger and needle nematodes infesting vineyards in southern Spain. European Journal of Plant Pathology, 129, 427–453. https://doi.org/10.1007/s10658-010-9705-y
Gutiérrez-Gutiérrez, C., Cantalapiedra-Navarrete, C., Decraemer, W., Vovlas, N., Prior, T., Palomares Rius, J. E., & Castillo, P. (2012). Phylogeny, diversity, and species delimitation in some species of the Xiphinema americanum-group complex (Nematoda: Longidoridae), as inferred from nuclear and mitochondrial DNA sequences and morphology. European Journal of Plant Pathology, 134, 561–597.
Gutiérrez-Gutiérrez, C., Cantalapiedra-Navarrete, C., Remesal, E., Palomares-Rius, J. E., Navas-Cortés, J. A., & Castillo, P. (2013). New insight into the identification and molecular phylogeny of dagger nematodes of the genus Xiphinema (Nematoda: Longidoridae) with description of two new species. Zoological Journal of the Linnean Society, 169(3), 548–579. https://doi.org/10.1111/zoj.12071
Gutiérrez-Gutiérrez, C., Bravo, M. A., Santos, M. T., Vieira, P., & Mota, M. (2016). An update on the genera Longidorus, Paralongidorus and Xiphinema (Family Longidoridae) in Portugal. Zootaxa, 4189(1), 099–114. https://doi.org/10.11646/zootaxa.4189.1.4
Gutiérrez-Gutiérrez, C., Mota, M., Castillo, P., Teixeira-Santos, M., & Palomares-Rius, J. E. (2018). Description and molecular phylogeny of one new and one known needle nematode of the genus Paralongidorus (Nematoda: Longidoridae) from grapevine in Portugal using integrative approach. European Journal of Plant Patholology, 151, 155–172. https://doi.org/10.1007/s10658-017-1364-9
Gutiérrez-Gutiérrez, C., Teixeira Santos, M., Inácio, M. L., Eisenback, J. D., & Mota, M. (2020). Description of Longidorus bordonensis sp. nov. from Portugal, with systematics and molecular phylogeny of the genus (Nematoda, Longidoridae). Zoosystematics and Evolution, 96(1), 175–193. https://doi.org/10.3897/zse.96.49022
Heyns, J., & Coomans, A. (1994). Four species of the Xiphinema americanum-group (Nematoda: Longidoridae) from islands in the western Indian Ocean. Nematologica, 40(1), 12–24.
Jahanshahi Afshar, F. (2019). The first molecular phylogenetic study of Xiphinema robbinsi Pedram, Niknam & Decraemer, 2008 (Nematoda: Longidoridae) based on specimens from the type locality. Nematology, 21(3), 333–335. https://doi.org/10.1163/15685411-00003219
Jahanshahi Afshar, F., Shahryari, F., Gharibzadeh, F., Pourjam, E., & Pedram, M. (2019). Description of Xiphinema azarbaijanense n. sp. (Nematoda; Longidoridae) from West Azarbaijan province, northwestern Iran. European Journal of Plant Patholology, 155, 417–434. https://doi.org/10.1007/s10658-019-01776-1
Jahanshahi Afshar, F., Pedram, P., & Mobasseri, M. (2021). Description of Xiphinema persicum n. sp. (Nematoda: Longidoridae), a X. americanum-group species from Iran. European Journal of Plant Pathology, 159, 783–797. https://doi.org/10.1007/s10658-021-02201-2
Jairajpuri, M. S., & Ahmad, W. (1992). Dorylaimida: Free-living, predaceous and plant-parasitic nematodes (p. 458). IBH Publishing Co.
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., Kikuchi, T., Manzanilla-Lopez, R., Palomares-Rius, J. E., Wesemael, W. M. L., & Perry, R. N. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology, 14, 946–961. https://doi.org/10.1111/mpp.12057
Katoh, K., Rozewicki, J., & Yamada, K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20(4), 1160–1166. https://doi.org/10.1093/bib/bbx108
Kawas, H. Z., & Al-Assas, K. M. (2011). Preliminary studies on viral diseases of olive in Damascus and Countryside and identification of accompanied nematodes. Journal of AL-Furat University, 272–293. [In Arabic].
Khaleel, H., Abu Al-Fadel, T., & Zeini, G. (2010). Survey and identification of parasitic nematode genera and species in the rhizosphere of apple trees at Homs governorate, Syria. Tishreen University Journal for Research and Scientific Studies - Biological Sciences Series, 32(4), 169–185. [In Arabic].
Kumari, S. (2006). Xiphinema simile (Nematoda: Longidoridae) in the Czech Republic and a note on other Xiphinema species. Helminthologia, 43, 43–50. https://doi.org/10.2478/s11687-006-0009-x
Kumari, S., Polák, J., & Choutka, R. (2005). Plant-parasitic nematodes of the genus Xiphinema (Nematoda: Longidoridae) in the vineyards of the Czech Republic. Nematology, 7(1), 81–93. https://doi.org/10.1163/1568541054192117
Lamberti, F. (1984). Nematode problems of the Mediterranean coastal stripe in the Syrian Arab Republic. Nematologia Mediterranea, 12, 53–64.
Lamberti, F., & Bleve-Zacheo, T. (1979). Studies on Xiphinema americanum sensu lato with descriptions of fifteen new species (Nematoda, Longidoridae). Nematologia Mediterranea, 7, 51–106.
Lamberti, F., & Siddiqi, M. R. (1977). Xiphinema pachtaicum [= X. mediterraneum]. CIH Descriptions of plant-parasitic. Nematodes Set, 7(94).
Lamberti, F., Choleva, B., & Agostinelli, A. (1983). Longidoridae from Bulgaria (Nematoda, Dorylaimida) with description of three new species of Longidorus and two new species of Xiphinema. Nematologia Mediterranea, 11, 49–72.
Lamberti, F., Ciancio, A., Agostinelli, A., & Coiro, M. I. (1991). Relationship between Xiphinema brevicolle and X. diffusum, with a redescription of Xiphinema brevicolle and description of three new species of Xiphinema (Nematoda, Dorylaimida). Nematologia Meditemanea, 19(3), 11–326.
Lamberti, F., Castillo, P., & Gómez-BarcinaAgostinelli, A. A. (1992). Descriptions of six new species of Xiphinema (Nematoda, Dorylaimida) from the Mediterranean region. Nematologia Mediterranea, 20, 125–139.
Lamberti, F., Lemos, R. M., Agostinelli, A., & D’addabo, T. (1993). The Xiphinema americanum-group in the vineyards of the Dao and Douro regions (Portugal) with description of two new species (Nematoda, Dorylaimida). Nematologia Mediterranea, 21(2), 215–225.
Lamberti, F., Molinari, S., De Luca, F., Agostinelli, A., & Di Vito, M. (1999a). Longidorids (Nematoda, Dorylaimida) from Syria with description of Longidorus pauli sp. n. and Paralongidorus halepensis sp. n. with SOD isozymes and PCR-RFLP profiles. Nematologia Mediterranea, 27, 63–78.
Lamberti, F., Sabova, M., De Luca, P., Molinari, S., Agostinelli, A., Coiro, M. I., & Valocka, B. (1999b). Phenotypic variations and genetic characterization of Xiphinema populations from Slovakia (Nematoda, Dorylaimida). Nematologia Meditemanea, 27(26), 1–275.
Lamberti, F., Molinari, S., Moens, M., & Brown, D. J. F. (2000). The Xiphinema americanum-group. I. Putative species, their geographical occurrence and distribution, and regional polytomous identification keys for the group. Russian Journal of Nematology, 8, 65–84.
Lamberti, F., Molinari, S., Moens, M., & Brown, D. J. F. (2002). The Xiphinema americanum-group. II. Morphometric relationships. Russian Journal of Nematology, 10, 99–112.
Lamberti, F., Hockland, S., Agostinelli, A., Moens, M., & Brown, D. J. F. (2004). The Xiphinema americanum group. 3. Keys to species identification. Nematologia Mediterranea, 32, 53–56.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, V., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., & Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 3(21), 2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Lazarova, S. S., Malloch, G., Oliveira, C. M., Hübschen, J., & Neilson, R. (2006). Ribosomal and mitochondrial DNA analyses of Xiphinema americanum-group populations. Journal of Nematology, 38(4), 404–410.
Lazarova, S. S., De Luca, F., & Peneva, V. K. (2008). On two closely related species of the Xiphinema americanum group: X. simile Lamberti, Choleva et Agostinelli, 1983 and X. parasimile Barsi et Lamberti, 2004 (Longidoridae), with a description of the male of X. parasimile. ZooKeys, 3, 29–50. https://doi.org/10.3897/zookeys.3.26
Lazarova, S. S., Brown, D. J., Oliveira, C. M., Fenton, B., MacKenzie, K., Wright, F., Malloch, G., & Neilson, R. (2016a). Diversity of endosymbiont bacteria associated with a non-filarial nematode group. Nematology, 18(5), 615–623. https://doi.org/10.1163/15685411-00002982
Lazarova, S., Peneva, V., & Kumari, S. (2016b). Morphological and molecular characterisation, and phylogenetic position of X. browni sp. n., X. penevi sp. n. and two known species of Xiphinema americanum-group (Nematoda, Longidoridae). ZooKeys, 574, 1–42. https://doi.org/10.3897/zookeys.574.8037
Lazarova, S., Oliveira, C. M., Prior, T., Peneva, V., & Kumari, S. (2019). An integrative approach to the study of Xiphinema brevicolle Lordello and Da Costa 1961, supports its limited distribution worldwide (Nematoda: Longidoridae). European Journal of Plant Pathology, 153(2), 441–464. https://doi.org/10.1007/s10658-018-1571-z
Lišková, M. (1995). Faunistic and ecological notes on Xiphinema (Nematoda: Dorylaimida) in Slovakia. Biológia (Bratislava), 50(2), 125–131.
Lišková, M., & Brown, D. J. F. (1996). Taxonomic validity and ecological relations of Xiphinema pachtaicum and X. simile (Nematoda: Dorylaimida), two members of the X. americanum group occurring in Slovakia. Helminthologia, 33, 137–142.
Lišková, M., Lamberti, F., Sabová, M., Valovská, B., & Agostinelli, A. (1993). First record of some species of longidorid nematodes from Slovakia. Nematologia Mediterranea, 21, 49–53.
Loof, P. A. A., & Coomans, A. (1968). On the development and location of the oesophageal gland nuclei in the Dorylaimina. Nematologica, 14, 596–597. https://doi.org/10.1163/187529268X00345
Loof, P. A. A., & Luc, M. (1990). A revised polytomous key for the identification of species of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) with exclusion of the X. americanum-group. Systematic Parasitology, 16, 35–66. https://doi.org/10.1007/BF00009600
Loof, P. A. A., & Luc, M. (1993). A revised polytomous key for the identification of species of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) with exclusion of the X. americanum-group. Systematic parasitology, 24, 185–189.
Loof, P. A. A., Luc, M., & Baujard, P. (1996). A revised polytomous key for the identification of species of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) with exclusion of the X. americanum-group: Supplement 2. Systematic Parasitology, 33, 23–29. https://doi.org/10.1007/BF00009717
Luc, M., Lima, M. B., Weischer, B., & Flegg, J. J. M. (1964). Xiphinema vuittenezi n. sp. (Nematoda: Dorylaimidae). Nematologica, 10, 151–163. https://doi.org/10.1163/187529264X00781
Luc, M., Coomans, A., Loof, P. A. A., & Baujard, P. (1998). The Xiphinema americanum-group (Nematoda: Longidoridae). 2. Observations on Xiphinema brevicollum Lordello & da Costa, 1961 and comments on the group. Fundamental and Applied Nematology, 21, 475–490.
Mobasseri, M., Hutchinson, M. C., Jahanshahi Afshar, F., & Pedram, M. (2019a). New evidence of nematode-endosymbiont bacteria coevolution based on one new and one known dagger nematode species of Xiphinema americanum-group (Nematoda, Longidoridae). PLoS ONE, 14(6), e0217506. https://doi.org/10.1371/journal.pone.0217506
Mobasseri, M., Hutchinson, M. C., Jahanshahi Afshar, F., & Pedram, M. (2019b). New evidence of nematodeendosymbiont bacteria coevolution based on one new and one known dagger nematode species of Xiphinema americanum-group (Nematoda, Longidoridae). PLoS ONE, 14(6), e0217506. https://doi.org/10.1371/journal.pone.0217506
Mojtahedi, H., Sturhan, D., Akhiani, A., & Barooti, S. (1980). Xiphinema species in Iranian vineyards. Nematologia Mediterranea, 8, 165–170.
Naghavi, A., Niknam, G., & Vazifeh, N. (2022a). A new species of Xiphinema americanum group (Nematoda: Longidoridae) from Iran, with additional data on three known species. Systematic Parasitology, 99(5), 545–555. https://doi.org/10.1007/s11230-022-10044-6
Naghavi, A., Niknam, G., & Fallahi, A. (2022b). Study of morphological, morphometric and molecular characteristics of two species Xiphinema vuittenezi Luc, Lima, Weischer &. Flegg, 1964 and X. primum Mobasseri, Hutchinson, Jahanshahi Afshar & Pedram, 2019. 24th Iranian Plant Protection Congress, 3rd-6th September, IRIPP, Tehran.
OEPP/EPPO. (2022). European and Mediterranean Plant Protection Organization. EPPO A1, A2 List of pests recommended for regulation as quarantine pests. (Last updated 2022/09).
Oliveira, C. M., Fenton, B., Malloch, G., Brown, D. J., & Neilson, R. (2005). Development of species-specific primers for the ectoparasitic nematode species Xiphinema brevicolle, X. diffusum, X. elongatum, X. ifacolum and X. longicaudatum (Nematoda: Longidoridae) based on ribosomal DNA sequences. Annals of Applied Biology, 146(3), 281–288. https://doi.org/10.1111/j.1744-7348.2005.040031.x
Orlando, V., Chitambar, J. J., Dong, K., Chizhov, V. N., Mollov, D., Bert, W., & Subbotin, S. A. (2016). Molecular and morphological characterisation of Xiphinema americanum-group species (Nematoda: Dorylaimida) from California, USA, and other regions, and co-evolution of bacteria from the genus Candidatus Xiphinematobacter with nematodes. Nematology, 18(9), 1015–1043. https://doi.org/10.1163/15685411-00003012
Palomares-Rius, J. E., Archidona-Yuste, A., Cantalapiedra-Navarrete, C., Prieto, P., & Castillo, P. (2016). Molecular diversity of bacterial endosymbionts associated with dagger nematodes of the genus Xiphinema (Nematoda: Longidoridae) reveals a high degree of phylogenetic congruence with their host. Molecular Ecology, 25(24), 6225–6247. https://doi.org/10.1111/mec.13904
Palomares-Rius, J. E., Cantalapiedra-Navarrete, C., Archidona-Yuste, A., Subbotin, S. A., & Castillo, P. (2017). The utility of mtDNA and rDNA for barcoding and phylogeny of plant-parasitic nematodes from Longidoridae (Nematoda, Enoplea). Scientific Reports, 7(1), 10905. https://doi.org/10.1038/s41598-017-11085-4
Palomares-Rius, J. E., Gutiérrez-Gutiérrez, C., Mota, M., Bert, W., Claeys, M., Yushin, V. V., Suzina, N. E., Ariskina, E. V., Evtushenko, L. I., Subbotin, S. A., & Castillo, P. (2021). ‘Candidatus Xiphinematincola pachtaicus’ gen. nov., sp. nov., an endosymbiotic bacterium associated with nematode species of the genus Xiphinema (Nematoda, Longidoridae). International Journal of Systematic and Evolutionary Microbiology, 71(7), 004888. https://doi.org/10.1099/ijsem.0.004888
Pearse, A. S. (1936). Zoological Names. A List of Phyla, Classes, and Orders Prepared for Section F. American Association for the Advancement of Science. Duke University Press. 24pp.
Pearse, A. S. (1942). Introduction to parasitology (p. 357). Springfield.
Pedram, M., Niknam, G., Robbins, R. T., Ye, W., & Karegar, A. (2009). Xiphinema iranicum n. sp. (Nematoda: Longidoridae) from north-western Iran. Nematology, 11, 11–21. https://doi.org/10.1163/156854108X398363
Pedram, M., Pourjam, E., Robbins, R. T., Ye, W., Atighi, M. R., & Decraemer, W. (2012). Morphological and molecular characterisation of Xiphinema mazandaranense n. sp. (Dorylaimida: Longidoridae), a new member of the Xiphinema pyrenaicum species complex. Nematology, 14, 109–119. https://doi.org/10.1163/138855411X580768
Peneva, V., & Choleva, B. (1992). Nematodes of the family Longidoridae from forest nurseries in Bulgaria. I. The genus Longidorus Micoletzky, 1922. Helminthology, 32, 35–45.
Potts, F. A. (1932). The phylum Nematoda. In L. A. Borradaile & F. A. Potts (Eds.), The invertebrata. A manual for the use of students (pp. 214–227). Macmillan.
Poureskandarian, M. A., Pourjam, E., & Pedram, M. (2023). New evidence on cryptic speciation in soil-inhabiting nematodes, with morphological and molecular characterisation of Xiphinema sangesarense n. sp. (Dorylaimida: Longidoridae) from north-central mountains of Iran. Nematology, 25(5), 549–563. https://doi.org/10.1163/15685411-bja10238
Rambaut, A. (2009). Figtree [Computer Software]. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 01 Sep 2023.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., & Suchard, M. A. (2018). Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67(5), 901–904. https://doi.org/10.1093/sysbio/syy032
Repasi, V., Agostinelli, A., Nagy, P., Coiro, M. I., Hecker, K., & Lamberti, F. (2008). Distribution and morphometrical characterization of Xiphinema pachtaicum, X. simile and X. brevicollum from Hungary. Helminthologia, 45, 96–102. https://doi.org/10.2478/s11687-008-0018-z
Robbins, R. T., Brown, D. J. F., Halbrendt, M. J., & Vrain, T. C. (1996). Compendium of juvenile stages of Xiphinema species (Nematoda: Longidoridae). Russian Journal of Nematology, 4, 163–171.
Roca, F., Lamberti, F., & Agostinelli, A. (1989). I Longidoridae (Nematoda, Dorylaimida) delle regioni Italiane. IX. La Sicilia. Nematologia Mediterranea, 17, 151–165.
Roca, F., Lamberti, F., & D’errico, F. P. (1991). I Longidoridae (Nematoda, Dorylaimida) delle regioni Italiane. XI. La Campania. Nematologia Mediterranea, 19, 139–154.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A., & Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
Seinhorst, J. W. (1959). A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica, 4, 67–69.
Shokoohi, E., Nasira, K., Iqbal, E., Hussain, S., & Mashela, W. P. (2021). First Report of Xiphinema diffusum from Pakistan. Helminthologia, 58(1), 92–99. https://doi.org/10.2478/helm-2021-0012
Tanabe, A. S. (2011). Kakusan4 and Aminosan: Two programs for comparing nonpartitioned, proportional, and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Molecular Ecolology Resources, 11(5), 914–921. https://doi.org/10.1111/j.1755-0998.2011.03021.x
Taylor, C. A., & Brown, D. J. F. (1997). Nematode-transmitted viruses. In F. Lamberti, C. E. Taylor, & J. W. Seinhorst (Eds.), Nematode vectors of plant viruses (pp. 142–143). CAB International.
Trudgill, D. L., Brown, D. J. F., & McNamara, D. G. (1983). Methods and criteria for assessing the transmission of plant viruses by longidorid nematodes. Revue De Nématologie, 6, 133–141.
Vazifeh, N., Niknam, G., Jabbari, H., & Naghavi, A. (2019). Description of a new dagger nematode, Xiphinema barooghii n. sp. (Nematoda: Longidoridae) and additional data on the three known species of the genus from northwest of Iran. Journal of Nematology, 51, 1–17. https://doi.org/10.21307/jofnem-2019-007
Walker, G. E. (2004). New Australian record for Xiphinema vuittenezi on Vitis vinifera. Australasian Plant Pathology, 33, 131–132. https://doi.org/10.1071/AP03071
Wang, S., Chiu, W. F., Yu, C., Li, C., & Robbins, R. T. (1996). The occurrence and geographical distribution of longidorid and trichodorid nematodes associated with vineyards and orchards in China. Russian Journal of Nematology, 4, 145–154.
Acknowledgements
The authors would like to acknowledge Inácia Ferreira and Teresa Rosmaninho (Universidade of Évora, Évora, Portugal) for their excellent technical assistance, and M. Hassan (Tishreen University, Latakia, Syria) for his critical reading and suggestions for this manuscript. The first author would like to thank A. Hasan and R. Ismail for their help in the soil sampling.
Funding
Open access funding provided by FCT|FCCN (b-on). This work is funded by National Funds through FCT—Foundation for Science and Technology under the Project UIDB/05183/2020 (Portugal) to MED (https://doi.org/10.54499/UIDB/05183/2020; https://doi.org/10.54499/UIDP/05183/2020) and CHANGE (https://doi.org/10.54499/LA/P/0121/2020).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Research involving human participants and/or animals
No specific permits were required for the described fieldwork studies. Permission for sampling the banana fields was granted by the landowner. However, the sites are not protected in any way.
Informed consent
All the authors certify that this work carried out in this research followed the principles of ethical and professional conduct have been followed. The funding source had no role the design of this study and not had any role during its execution, data collection, analyses, interpretation of the data, decision to publish, or preparation of the manuscript.
Conflict of interest
All authors declare no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ali, N., Vicente, C.S.L., Mota, M. et al. First report of four dagger nematode species of the genus Xiphinema (Nematoda: Longidoridae) from banana in Syria using an integrative approach. Eur J Plant Pathol (2024). https://doi.org/10.1007/s10658-024-02868-3
Accepted:
Published:
DOI: https://doi.org/10.1007/s10658-024-02868-3